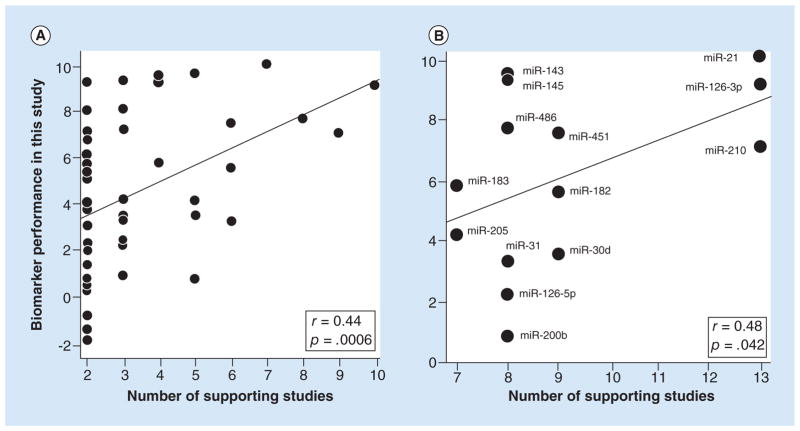
Figure 1. Scatter plots showing that number of supporting studies predicted the performance of biomarker candidates.
The performance values (y-axis) were derived by rescaling the differential expression p-values in Tables 1 & 2 and using negative values for three of the miRNAs because their expression in tumor versus normal was opposite from the direction expected (see statistics methods). The lines are the best fit lines from the Pearson correlation analysis, the r’s are correlation coefficients and the p’s are the significance values of the correlations (1-tailed). (A) Correlation with the number of supporting studies determined by Guan et al. [2]. n = 50 miRNA biomarkers. (B) Correlation with the number of supporting studies determined by Vosa et al. [3]. n = 14 miRNA biomarkers.