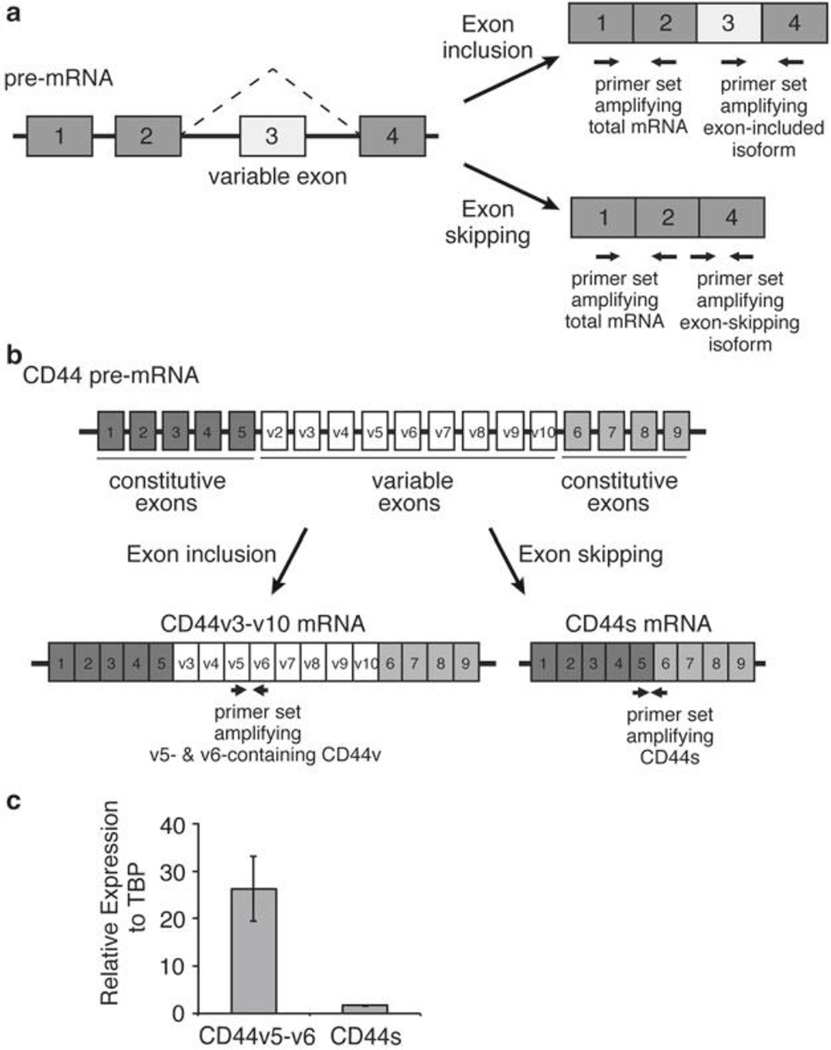
Fig. 1.
Characterization of alternative splicing by qRT-PCR. (a) A schematic illustrating a hypothetical four-exon gene with a variable exon 3 that undergoes exon-inclusion or exon-skipping. Three primer sets for qPCR indicated by the paired arrows are designed to amplify the total mRNA, the variable exon 3 inclusion isoform, or the exon-skipping isoform. (b) A schematic depicting the exon structure of the transmembrane protein CD44, with the nine constitutive and nine variable exons labeled. CD44v3-v10 and CD44s are predominant splice isoforms and are depicted. Paired arrows indicate primer sets to amplify CD44v v5-v6 containing isoforms and the CD44s isoform which lacks all variable exons. (c) Representative qRT-PCR results using primers in Table 1 to show the relative expression of CD44v5/v6 and CD44s, normalized to TBP, in an immortalized human mammary epithelial cell line (HMLE)