Figure 1. Key traits and phylogenetic relationships of the 86 yeasts of the subphylum Saccharomycotina whose genomes have been sequenced.
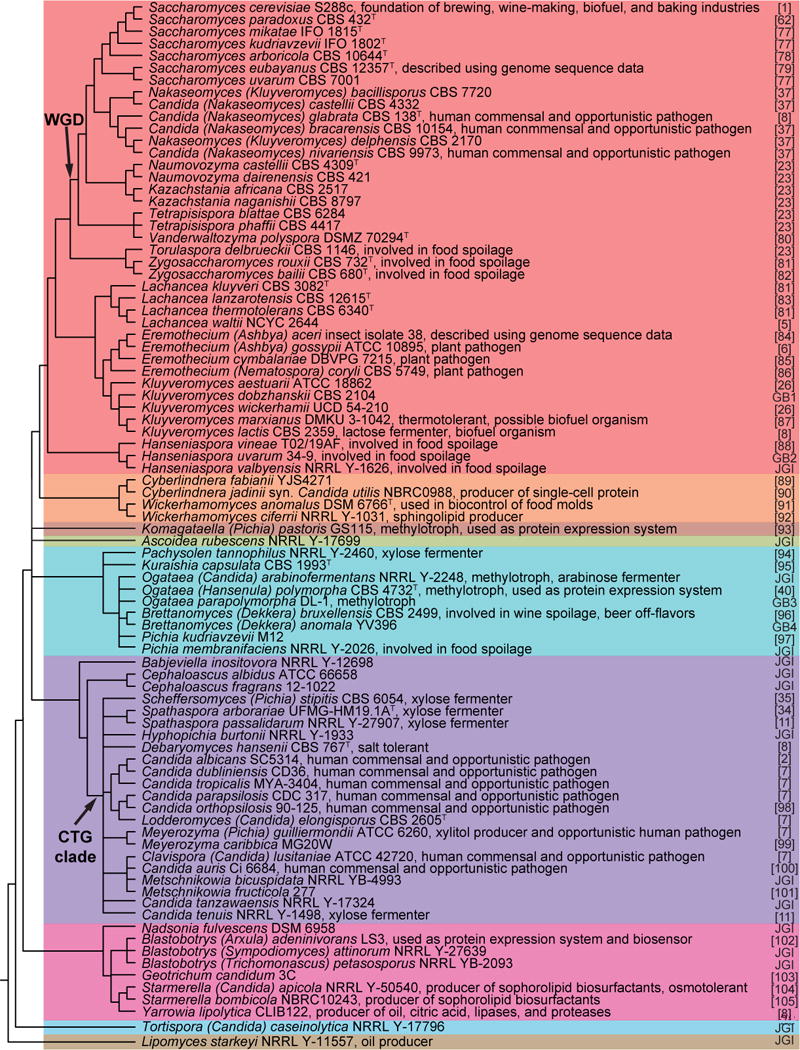
The topology of the cladogram has been estimated conservatively from previous analyses using genome [12,23] or multi-locus sequence data [27,36]. Major clades [36] are color-coded (clade names are shown in Figure 2). Only one reference genome per species is included with preference given to the highest quality and/or most widely used reference genome. Only publicly available genome assemblies are included. Interspecies hybrids are discussed in the text but are not shown here. WGD, whole genome duplication; recent work has shown that the WGD was caused by an allopolyploidization event that occurred between an early member of the Zygosaccharomyces/Torulaspora clade and an early member of the Kluyveromyces/Lachancea/Eremothecium clade, thus making this part of the phylogeny a network, rather than a tree [73]. CTG clade, yeasts using an alternate codon table where CTG encodes serine, instead of leucine. JGI, genomes publicly available on MycoCosm at http://genome.jgi-psf.org/programs/fungi/index.jsf, which are subject to the usage terms of the DOE Joint Genome Institute until formal publication. GB1, GenBank Accession CCBQ000000000; GB2, Genbank Accession JPPO00000000; GB3, GenBank Accession AEOI00000000; GB4, GenBank Accession LCTY00000000.
