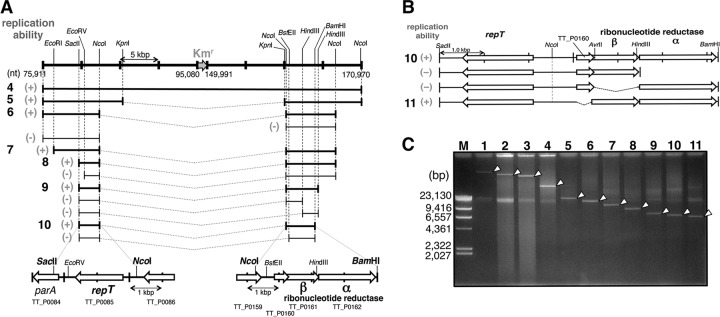
FIG 2.
Minimization of pTT27 in a stepwise manner. (A) Scheme for the stepwise deletions. The plasmids were constructed in E. coli, and replication ability was confirmed by transformation of the recA-null mutant of HB27. The top line indicates the pUC27H4 from Fig. 1B, panel i, and the numbers are positions on the intact pTT27. The lower lines represent the residual region, and their replication abilities are indicated in a manner similar to that in Fig. 1B. The V shape of the dotted line indicates a linkage between the two ends. In the bottom image, the two pTT27 regions included in pUC27H10 (of 10) are schematically shown. (B) Involvement of three genes, TT_P0160, TT_P0161 (RNR-β), and TT_P0162 (RNR-α), in pTT27 replication. Deletion of TT_P0161 or TT_P0162, which would compose the class I RNR enzyme, resulted in no transformant of the recA-null mutant, having a transformation efficiency below 1.0 × 100 CFU/μg. (C) Stepwise deletion in pTT27 shown by normal agarose gel electrophoresis of the pTT27 derivatives extracted from HB27 without digestion. The arrowheads indicate the pTT27 derivatives. Lane M, size marker of HindIII-digested λ DNA. The other lane numbers correspond to the leftmost boldface ones in Fig. 1B (panel i) and 2A. In the small-scale plasmid preparation from T. thermophilus by the alkali-SDS method, the shattered genomic DNA is often observed, at around 20 kbp.