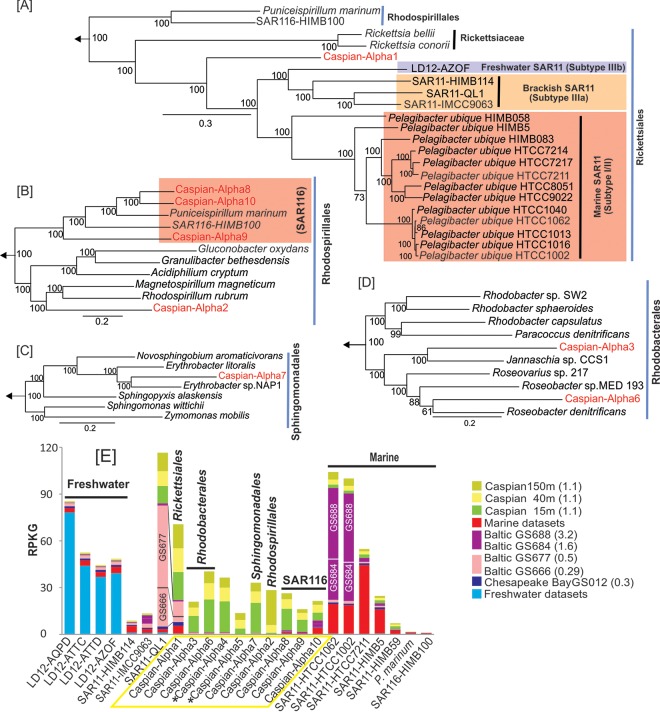
FIG 3.
Phylogeny of the genomes belonging to the phylum Alphaproteobacteria in the Caspian Sea and their metagenomic recruitments across diverse aquatic habitats. (A to D) Maximum likelihood phylogenies, obtained using a concatenation of conserved proteins, of Caspian Sea genomes and their close relatives in the orders Rickettsiales (190 proteins) (A), Rhodospirillales (377 proteins) (B), Sphingomonadales (167 proteins) (C), and Rhodobacterales (D). Genomes from the class Gammaproteobacteria were used to root these trees. The Caspian Sea genomes are shown in red. Bootstrap values (in percent) are indicated at each node. (E) Metagenomic recruitment of Caspian Sea alphaproteobacterial genomes and related reference genomes in different data sets. The data sets used are described in Fig. 1. (Left) Genomes of organisms of freshwater origin; (middle) genomes of organisms of brackish water origin; (right) genomes of organisms of marine water origin. The salinity of the samples is indicated in parentheses. An asterisk next to a Caspian Sea genome indicates a small genome (<500 kb) that could be classified only as belonging to the Alphaproteobacteria and no further.