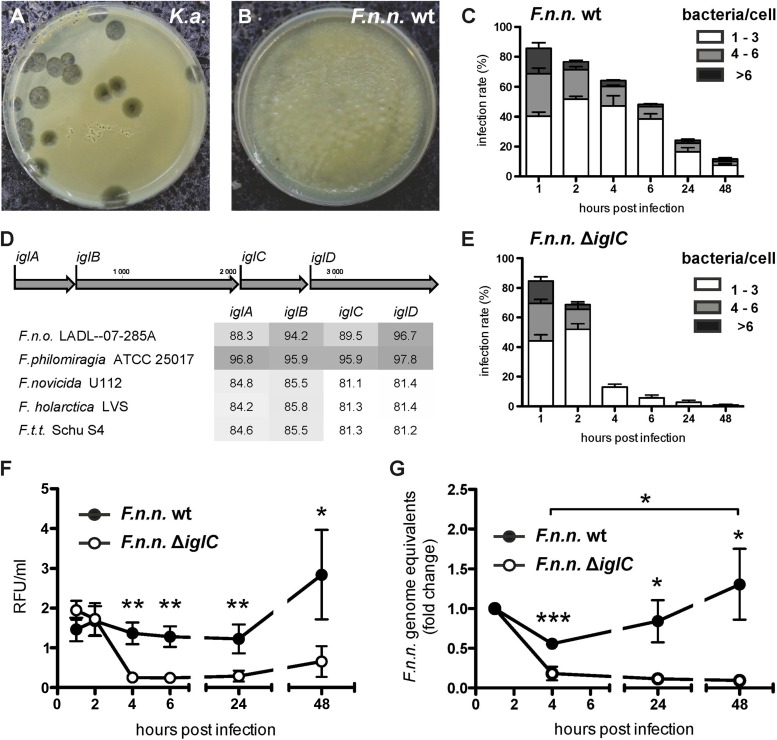
FIG 1.
Persistence and replication of F. noatunensis subsp. noatunensis in Dictyostelium is dependent on IglC. (A and B) Plaque assay to determine resistance of bacteria to feeding of Dictyostelium. Dictyostelium feeds and forms plaques on a lawn of K. aerogenes (K.a.) (A), but not on F. noatunensis subsp. noatunensis (F.n.n.) wt (B). Shown are images of one representative experiment (n = 3). (C and E) Infection rate (y axis) and bacteria/cell (bar sections) of F. noatunensis subsp. noatunensis wt (C) and ΔiglC (E) in Dictyostelium wt cells over 48 hpi. Data were collected from 7 replicates, and a minimum of 100 cells were counted. The error bars indicate standard errors of the mean (SEM). (D) Nucleotide sequence comparison of iglA to iglD in F. noatunensis subsp. noatunensis FSC769 with other selected Francisella spp. (F.t.t., F. tularensis subsp. tularensis; F.n.o., F. noatunensis subsp. orientalis). The results are presented as percent nucleotide identity with the iglA to iglD loci of F. noatunensis subsp. noatunensis FSC769 after pairwise comparison. The depth of gray shading indicates the level of sequence identity, with dark gray representing the highest similarity. (F and G) Bacterial replication of F. noatunensis subsp. noatunensis wt and ΔiglC was measured by flow cytometry analysis of bacterial green fluorescence per unit volume of infected cell culture (F) and by qPCR of F. noatunensis subsp. noatunensis genomes normalized to host cell genomes (G). Data from 8 (F) and 5 (G) independent experiments (means ± SEM) were collected (unpaired, two-tailed t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).