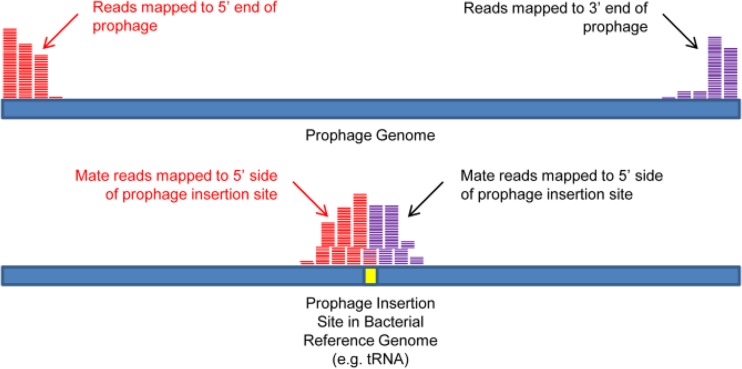
FIG 3.
End-sequence profiling for prophage insertion site detection. Paired-end reads are aligned to a multifasta reference containing a phage sequence-free bacterial genome and multiple phage genomes. When the ends of an index phage have homology to one of the sequenced strain's phages, one read of the pair will map to the index phage and the other will map to the corresponding side of the phage insertion site in the bacterial genome. Opposite ends of the phage will have mate reads mapped to different sides of the insertion site. Thus, the orientation of the phage can also be determined.