FIG 4.
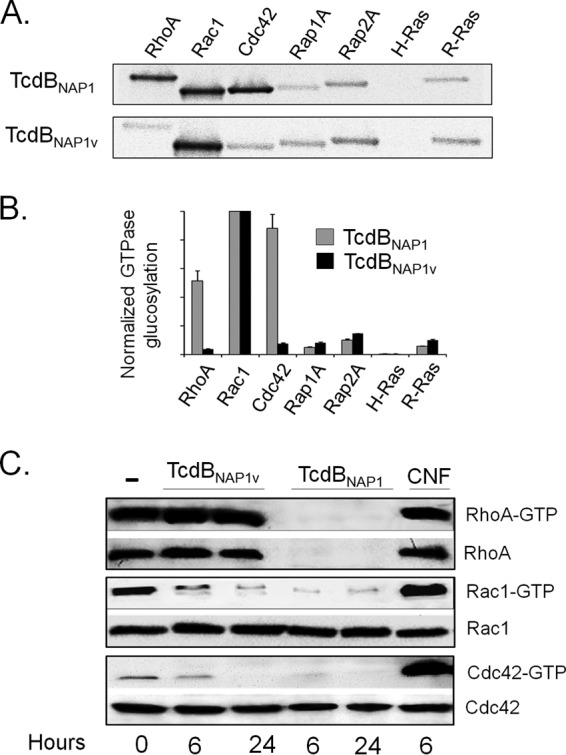
The NAP1V strain does not glucosylate RhoA. (A) TcdBNAP1 and TcdBNAP1V were tested for their ability to glycosylate a panel of recombinant GTPases using UDP-[14C]glucose as a cosubstrate. Labeled bands were detected by phosphorimaging analysis. (B) The band intensities of the GTPase glycosylation were quantified by densitometry. Each experiment was normalized to Rac1 signal. Means ± standard deviations from three independent experiments are shown. (C) Effect of TcdBNAP1 and TcdBNAP1V on the activation state of small GTPases. 3T3 fibroblasts were intoxicated with TcdBNAP1 and TcdBNAP1V for the indicated times. After treatment, cells were lysed. One part of the lysates was used as a control for total amount of GTPases, and the other one was incubated with PBD-GST or RBD-GST-Sepharose beads. Active proteins were pulled down and analyzed by Western blotting. GTPases were detected using anti-RhoA, anti-Rac, and anti-Cdc42, respectively. Cells treated with TcdBNAP1 show inactivation of RhoA, whereas cells intoxicated with TcdBNAP1V do not. Cytotoxic necrotizing factor 1 (CNF) from Escherichia coli was used as a positive control for GTPase activation. Negative-control cells were left untreated.
