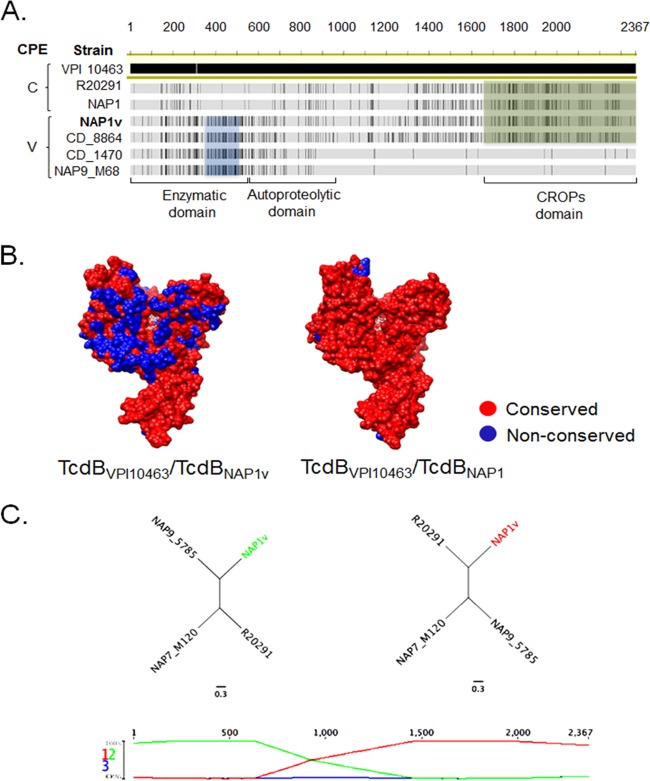
FIG 8.
TcdBNAP1V shares with TcdBNAP1 the receptor-binding domain but not the enzymatic domain. (A) Sequence alignment of (i) variant toxins B from NAP9_M68, CD_1470, and CD_8864 reference strains inducing a variant (V) CPE; (ii) TcdBNAP1 from a clinical isolate and a reference epidemic NAP1/RT027 strain (R20291) inducing a classic (C) CPE; and (iii) TcdBNAP1V. Black lines represent disagreements in the sequence of TcdBVPI10463, which was selected as a reference for the alignment. The blue box highlights a distinct glucosyltransferase region shared between NAP1V and other variant strains. The green box shows sequence stretches in the repetitive CROPs domains shared between TcdBNAP1, TcdBR20291, and TcdBNAP1V. (B) Comparison of the TcdBNAP1 and TcdBNAP1V sequences in the context of the TcdB GTD structure (PDB 2BVM, VPI 10463 sequence). Sequence conservation on the putative GTPase-binding face compared to the GTD from C. difficile VPI 10463 with NAP1 and NAP1V TcdB GTD structures is shown (red, conserved; blue, not conserved). UDP-glucose is depicted in white in the GTD active site. (C) Resulting recombination detection graphs using TcdB sequences from strains NAP9 (M68, RT017 reference strain), R20291 (epidemic NAP1/RT027 reference strain), NAP7 (epidemic M120, RT078 reference strain), and NAP1V. Signs of possible recombination events are represented as changes in the topology graphs (first row) that appear at the most probable topology between the segments. The cross of the topological lines (green and red lines) indicates recombination breakpoints. Resulting trees are compatible with a scenario in which TcdBNAP1V emerged through recombination of tcdB sequences from NAP9 and NAP1 strains.