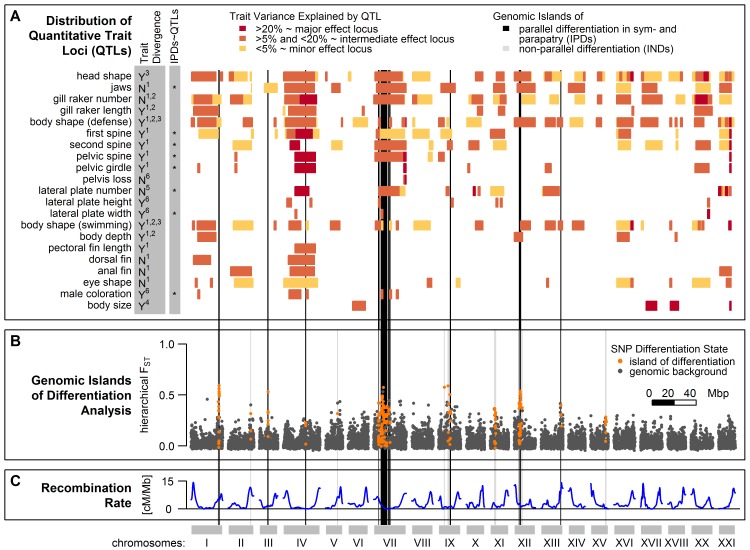
Fig 4. Genomic islands of differentiation among Lake Constance stickleback and distribution of Quantitative Trait Loci (QTL).
(B) Of 37 genomic islands of differentiation identified in Lake Constance stickleback, 19 showed parallel differentiation between lake and stream ecotypes both in sympatry and parapatry (IPDs, black vertical bars), while non-parallel differentiation in 18 further islands (INDs, grey vertical bars) were mainly driven by differentiation between the parapatric ecotype comparison only. Dots show SNPs assigned to genomic islands of differentiation (orange) or the neutral genomic background (dark grey). (A) QTLs for traits previously studied among Lake Constance ecotypes and their overlap with parallel islands are shown. The left grey column indicates if traits have previously been found to be divergent among Lake Constance ecotypes (‘Y’ = yes) or not (‘N’ = no). Significant clustering of parallel islands inside QTLs for trait groups are indicated by asterisks in the right grey column. Blocks indicate 95% QTL confidence intervals (extent along x-axis) and effect sizes (color). References for phenotypic data: 1[59], 2[57], 3[65], 4[56] and S7B Fig, 5[46], 6S7A Fig. (C) Recombination rates across the stickleback genome estimated by Roesti et al. [77].