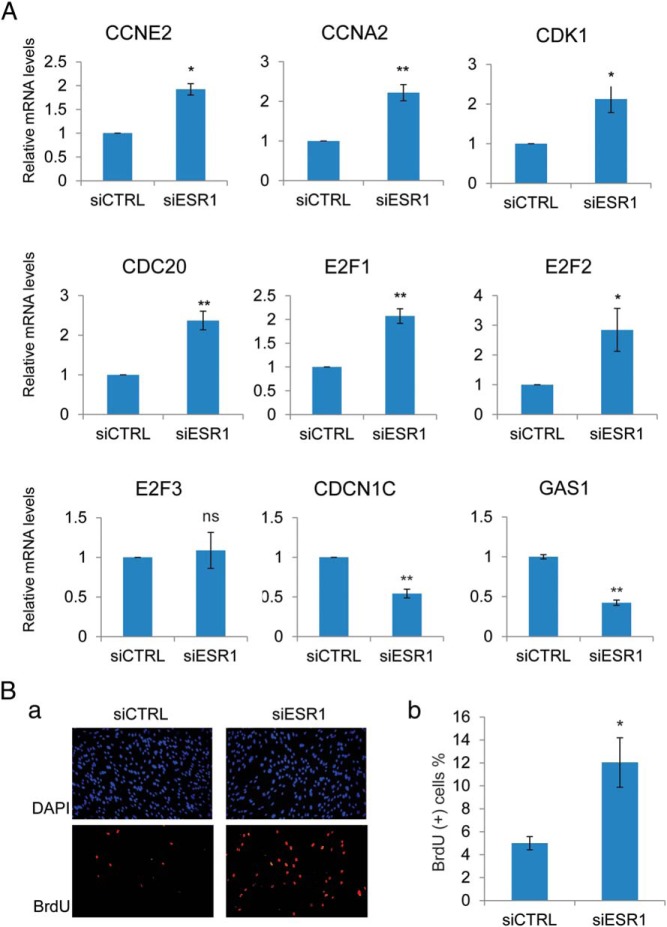
Figure 2.
A, HESCs were transfected with siRNA targeting ESR1 or scrambled siRNA (control [siCTRL]). Forty-eight hours after transfection, HESCs were treated with a differentiation cocktail to initiate in vitro differentiation. Cells were harvested at 24 hours after a differentiation cocktail treatment. Total RNA was isolated and cDNAs were prepared. cDNAs were subjected to real-time PCR using gene-specific primers for 36B4, CCNE2, CCNA2, CDK1, CDC20, E2F1, E2F2, E2F3, CDKN1C, and GAS1. PCR data are normalized to 36B4 mRNA levels. Relative fold changes were normalized with respect to control nontargeting siRNA treatment. Mean ± SEM of at least three independent experiments are shown. The Student's t test was used for statistical analysis. **, P ≤ .01; *, P ≤ .05. E, HESCs were synchronized at the G1/S stage by double-thymidine block and transfected with siRNA targeting ESR1 or scrambled siRNA (control). Twenty-four hours after transfection, HESCs were treated with differentiation cocktail to initiate in vitro differentiation. At 18 hours after a differentiation cocktail treatment, BrdU was added to the medium (100 μM). At 6 hours after BrdU addition (24 h of differentiation), cells were fixed and subjected to immunofluorescence to detect BrdU-positive cells, which are indicated in red. 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei are shown in blue. a, Images are the representatives of three independent experiments. b, The graph shows the mean values of percentages of BrdU-positive cells calculated from five fields in three different experiments. The Student's t test was used for statistical analysis. *, P ≤ .05.