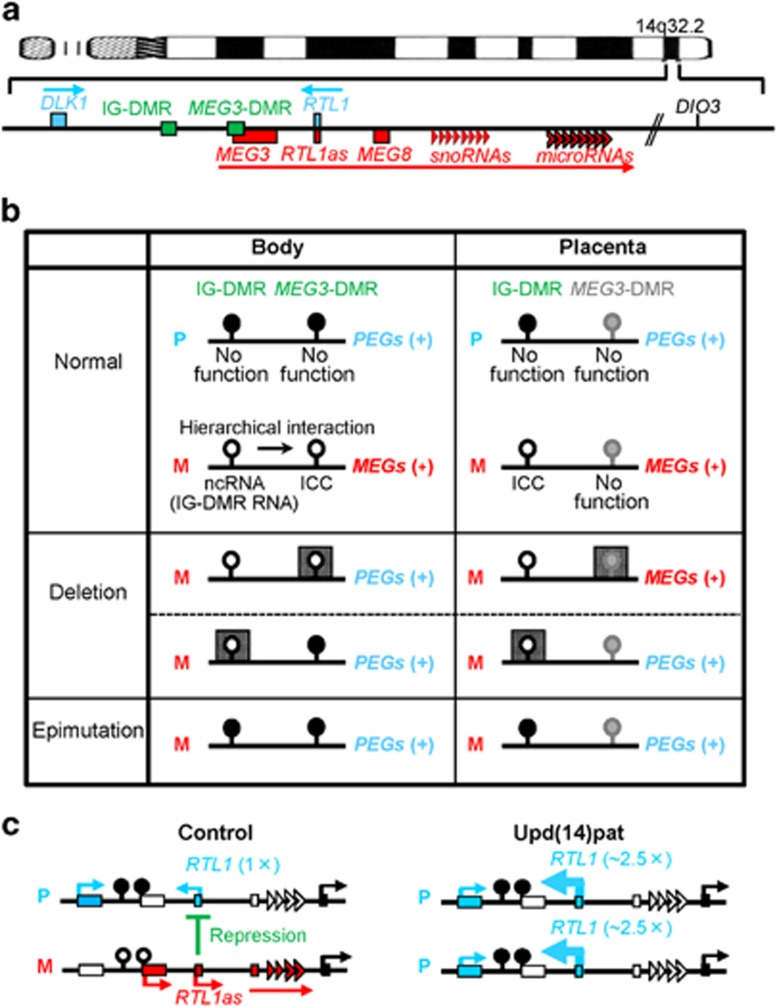
Figure 2.
The human chromosome 14q32.2 imprinted region. (a) Schematic representation of the physical map of this region. PEGs are shown in blue, MEGs in red, a probably non-imprinted gene (DIO3) in black and the DMRs in green. (b) Methylation patterns of the DMRs. Black, white and gray painted circles represent methylated DMRs, unmethylated DMRs and non-DMRs, respectively. The arrow indicates a hierarchical interaction between the IG-DMR and the MEG3-DMR that appears to be medicated by cis-acting ncRNAs (the IG-DMR RNA) that exerts an enhancer-like function for the MEG3 promoter and prevents the MEG3-DMR from methylation. The deleted regions are shown with stippled squares. (c) Interaction between RTL1 and RTL1as. In control subjects, RTL1as-encoded microRNAs function as a trans-acting repressor for RTL1. In upd(14)pat patients, RTL1 expression level becomes ~5 times increased because of two copies of functional RTL1 and no functional RTL1as (shown with thick arrows). DMR, differentially methylated region; ICC, imprinting control center; IG-DMR, intergenic differentially methylated region; M, maternally derived chromosome; MEGs, maternally expressed genes; ncRNA, noncoding RNA; P, paternally derived chromosome; PEGs, paternally expressed genes; upd(14)pat, paternal uniparental disomy 14.