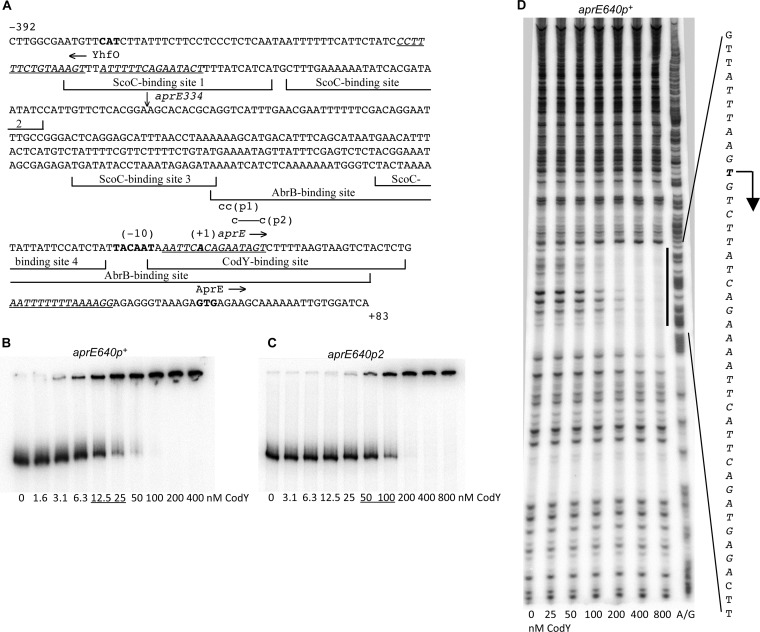
FIG 1.
Binding of CodY to the aprE regulatory region. (A) Sequence (5′ to 3′) of the coding (nontemplate) strand of the aprE regulatory region within the aprE640p+-lacZ fusion. Coordinates are reported with respect to the transcription start point (10). The upstream endpoints of inserts within the aprE640 and aprE334 fusions are at positions −557 and −251, respectively; the latter junction is indicated by a vertical arrow above the sequence. The downstream endpoints of both inserts coincide with the 3′ end of the presented sequence. The likely translation initiation codon, the −10 promoter region, and the apparent transcription start point are shown in bold. The directions of transcription and translation are indicated by horizontal arrows. The sequences that were protected by CodY (this work), ScoC (12), or AbrB (11) in DNase I footprinting experiments are shown by bracketed lines. The sequences of the four CodY-binding motifs, with three or four mismatches each, are italicized and underlined. The mutated nucleotides are shown in lowercase above the sequence. (B and C) Gel shift assays of CodY binding to aprE fragments. The aprE640p+ (B) and aprE640p2 (C) PCR fragments obtained with oligonucleotides oBB67 and oBB102, using pGB1 and pGB13, respectively, as templates, and labeled on the template strand were incubated with increasing amounts of purified CodY in the presence of 10 mM ILV. CodY concentrations used (monomers) are reported below the lanes; concentrations corresponding to the apparent KD for binding are underlined. (D) DNase I footprinting analysis of CodY binding to the aprE regulatory region. The aprE640p+ PCR fragment used for panel B was incubated with increasing amounts of purified CodY in the presence of 10 mM ILV and then with DNase I. The protected area is indicated by a vertical line, and the corresponding sequence is reported; the protected nucleotides are italicized. The apparent transcription start point and direction of transcription are shown by a bent arrow. CodY concentrations used (nanomolar [monomers]) are indicated below the lanes. The A+G sequencing ladder of the template DNA strand is shown in the right lane.