Figure EV1. Spindly, CLIP‐170, and CENP‐E are required for chromosome alignment, but not for cortical dynein recruitment.
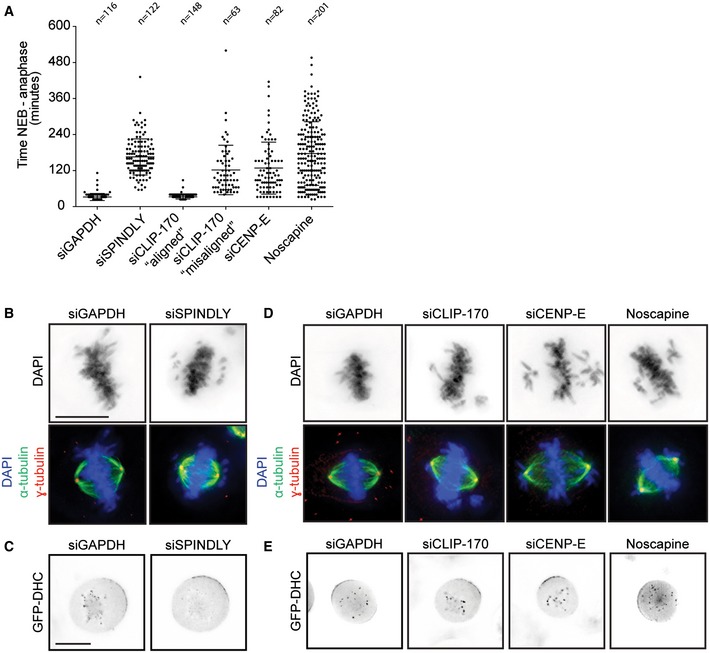
- Average time in mitosis (nuclear envelope breakdown till anaphase) of U2OS cells transfected with GAPDH, Spindly, CLIP‐170, and CENP‐E siRNA or treated with noscapine. Data adapted from experiments shown in Fig 1. Graph shows mean ± SD. (B, C) Scale bars represent 10 μm.
- Representative image of the mitotic phenotype observed after Spindly depletion. Cells were stained with α‐tubulin, γ‐tubulin, and DAPI.
- Representative live‐cell microscopic images of HeLa cells stably expressing GFP‐DHC from a bacterial artificial chromosome [49]. Cells were transfected with the indicated siRNAs for 48 h. One hour before images were taken, cells were treated with the Eg5 inhibitor STLC to arrest them in mitosis with a monopolar spindle and to allow the visualization of KT‐ and cortical dynein. Scale bar represents 10 μm.
- Cells were transfected with CLIP‐170 or CENP‐E siRNA for 48 h or treated for 30 min with noscapine and fixed and stained as in (B).
- Same experiment as described in (C) to examine GFP‐DHC localization after CLIP‐170 and CENP‐E depletion and noscapine treatment.
