Figure 3. Ephemeral life of PINK1 in the healthy mitochondria.
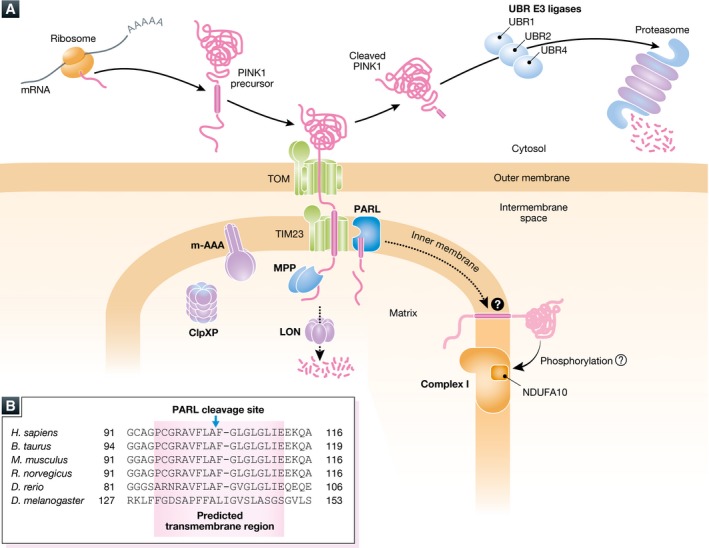
(A) The newly synthesized PINK1 precursor on the cytosolic ribosomes is targeted to the mitochondria. After crossing the outer membrane through the TOM complex, the N‐terminal mitochondrial targeting sequence is cleaved by MPP in the matrix. The following transmembrane segment is recognized by the TIM23 complex and received second cleavage by PARL between A103 and F104. Most of the cleaved PINK1 is released to the cytosol where the newly N‐terminal phenylalanine residue of the cleaved PINK1 is recognized by the N‐end rule UBR ligases (UBR1, UBR2, and UBR4 that preferentially recognize type‐2 N‐degrons) for proteasomal degradation. A matrix ATPase associated with diverse cellular activities (m‐AAA) protease is composed of AFG3L2 and paraplegin and has the active site oriented toward the matrix. ClpXP is a matrix ATP‐dependent protease composed of hetero‐oligomeric ATP‐binding subunits and proteolytic subunits. m‐AAA and ClpXP also participate in PINK1 degradation. Another ATP‐dependent LON protease also contributes, particularly in Drosophila, to PINK1 degradation in the matrix. As phosphorylation of NDUFA10 in the respiratory chain complex I is reduced in Pink1‐KO mice, a small amount of PINK1 retained in the inner membrane might be involved in the maintenance of complex I through phosphorylation. (B) Amino acid sequence alignment of the transmembrane region of PINK1. Amino acid sequences of the predicted transmembrane segments (pink‐colored box) from the indicated species are shown 99. The transmembrane regions are well conserved from zebrafish (Danio rerio) and humans (Homo sapiens), while the transmembrane segment in fly (Drosophila melanogaster) is less conserved with fewer glycine/proline residues. The PARL cleavage site of human PINK1 between A103 and F104 is also shown.
