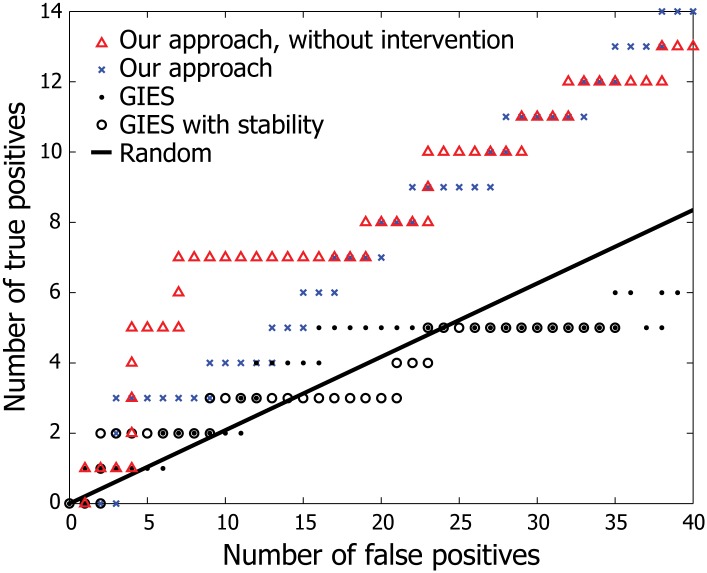
Fig 2. Reconstruction performance on single cell gene expression data.
We applied our Bayesian structure learning algorithm based on GBNs to uncover the signaling pathway of 11 human proteins from expression data provided by Sachs et al. [5]. MAP estimates of edge weights calculated using 1,000 posterior graph samples are used to generate a ranked list of (directed) edges for evaluation of accuracy. The data points for GIES are taken from Hauser and Bühlmann [19] for comparison. The result suggests GBNs can uncover causal edges in real biological networks, and that our approach is more effective than GIES.