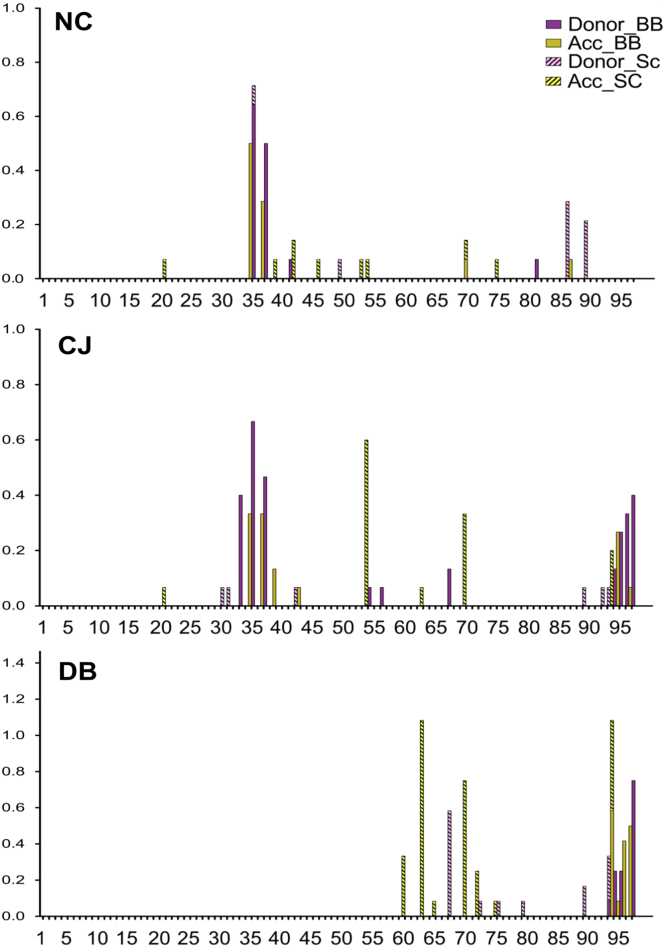
Fig. 8.
Inter-molecular hydrogen bond sites on SUMO. We detected hydrogen bonds using Rosetta hydrogen bond score. Each hydrogen bond was classified as either a donor or acceptor and if the chemical moiety participating in the hydrogen bond belongs to the peptide backbone or the side chain. The SUMO structures are grouped by the following protein–protein interactions: NC: noncovalent, CJ: conjugated, DB: deubiquitinase. The Y-axis represents average number of hydrogen bonds per PDB. The SUMO1 numbering scheme is used for all SUMO molecules (Fig. 1). Redundant hydrogen bonds in structure containing multiple SUMO chains were only counted once per PDB.