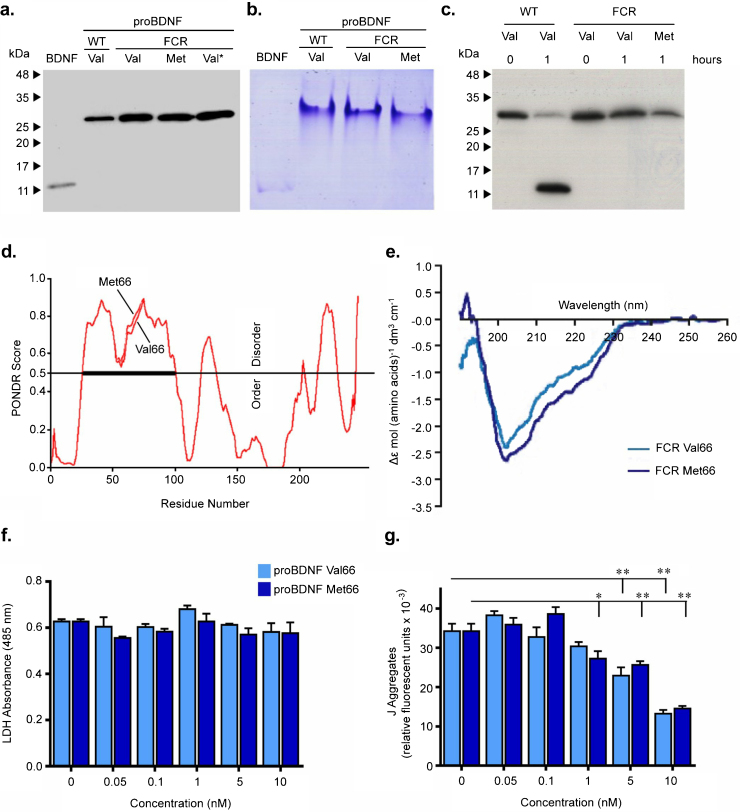
Fig. 1.
Characterization of proBDNF Val66Met FCR polymorphic variants. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
(a) Wild-type (WT) proBDNF or furin cleavage-resistant (FCR) proBDNF Val66Met variants (50 ng) and BDNF (10 ng) resolved with SDS PAGE and western blotted using anti-BDNF-N20 1:3000. Val* denotes commercial proBDNF Val66-FCR (Alomone labs). ProBDNF variants have a similar molecular weight of ∼26 kDa (unglycosylated) with high purity. (b) ProBDNF (∼1 μg) resolved on acidic native PAGE gels stained with Coomassie Blue showed no laddering or unresolved proteins, confirming absence of oligomerization and soluble aggregates. Mature BDNF (∼14 kDa) used as standard. (c) ProBDNF variants (50 μg/ml) furin digested 1 h at 30 °C, resolved on 12% SDS PAGE and analyzed using western blotting. Protein bands were detected using anti-BDNF N20 at 1:3000. WT was the positive control, cleaved to mature BDNF. Cleavage-resistant proBDNF variants were intact following furin exposure. (d) Overlay of PONDR outputs of Val66 and Met66. Values are predicted and scaled 0–1. The thick black line (‘disorder bar’) denotes strength of disorder prediction. (e) ProBDNF variants (∼0.1 mg/ml) were measured by CD in the far-UV range. Spectra of both variants showed predominant β-structure with subtle differences in the 222 nm region. Lines show Val66 in light blue Met66 in dark blue. (f) and (g) E16 cortical neurons (DIV11) were serum starved for 2 h and treated with a range of concentrations (0–10 nM) of proBDNF variants for 18 h, after which cells were processed and measured at (f) 485 nm for LDH release (ProBDNF FCR Val66 in light blue, Met66 in dark blue) or (g) at excitation/emission 560/595 nm for JC-1 assay with relative fluorescence of J-aggregates measured (ProBDNF WT Val66 in light blue, Met66 in dark blue). The polymorphism was not associated (P = 0.473) with the reduction in membrane potential (two-way ANOVA followed by Bonferroni’s post hoc test). In (f) and (g) changes are represented as column graphs with subsequent analysis using one-way ANOVA followed by Dunnett’s multiple comparison post hoc test for comparison within group. Throughout, error bars denote standard error of mean. Significance is denoted as *P < 0.05, **P < 0.01 and ***P < 0.001.