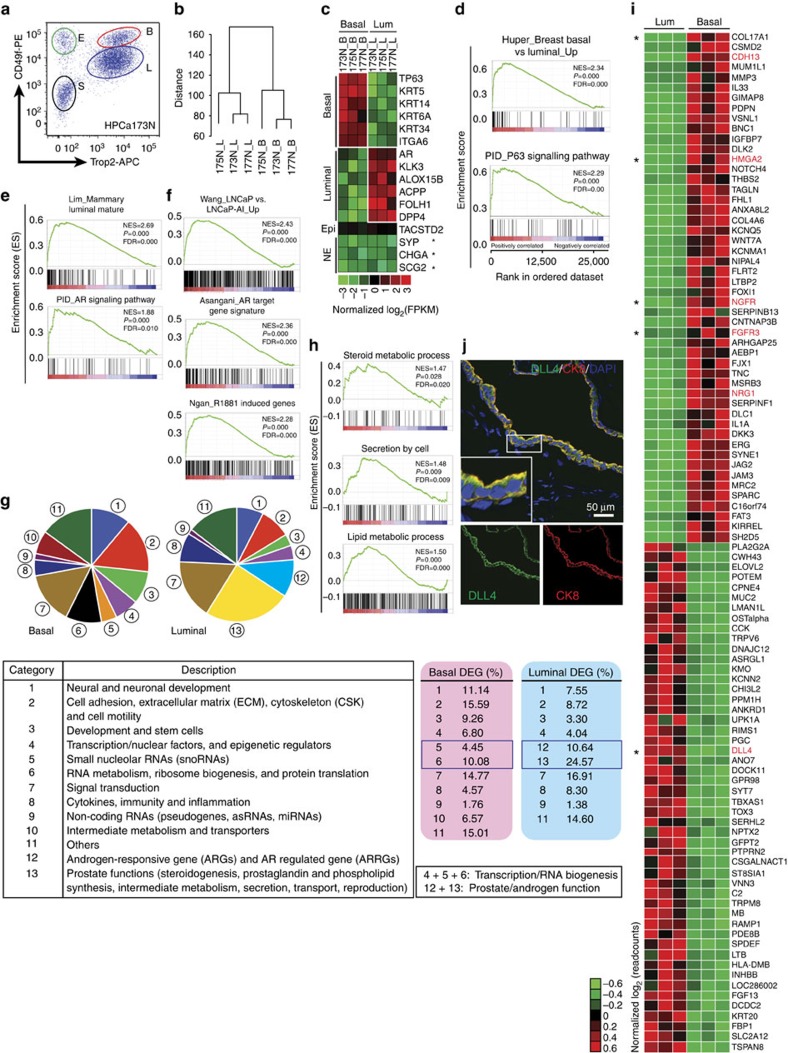
Figure 1. Distinct gene-expression profiles of prostatic basal and luminal cells.
(a) FACS plots of prostate basal (B), luminal (L), endothelial-enriched (E) and stromal-enriched (S) populations identified as Trop2+CD49fhi, Trop2+CD49flo, Trop2-cd49fhi and Trop2-CD49f-, respectively. (b) Hierarchical clustering of RNA-Seq data in three pairs of benign prostatic basal (N_B) and luminal (N_L) cells. All detected genes were used in the clustering analysis. y-axis shows euclidean distance for log2 (normalized read counts). (c) Heatmap presentation of expression of known phenotypic markers corresponding to different prostate cell lineages. Asterisks indicates the genes expressed at very low levels (FPKM<0.18). (d–f) Representative GSEA results in basal (d) and luminal (e) cells. In (f), androgen-responsive genes and AR-regulated genes in the three indicated data sets are enriched in luminal cells. (g) Distinct transcriptomic profiles of human prostatic basal and luminal cells. Shown are pie charts of gene categories (Supplementary Data 1) over-represented in basal and luminal populations. Descriptions of each functional category are given in the table below. Percentages of each category in basal versus luminal cells are marked in red and blue, respectively (below, right). (h) GSEA results for the enrichment of indicated gene signatures in luminal cells. (i) Heatmap of the top 50 putative marker genes in each epithelial lineage. Asterisks indicate the genes whose exclusive cellular localization has been confirmed by immunofluorescence. Genes with their biological functions investigated in this study are coloured in red. See the ‘Methods' section for detail. (j) Immunofluorescence of DLL4 and CK8 in benign prostate tissues. Scale bars, 50 μm.