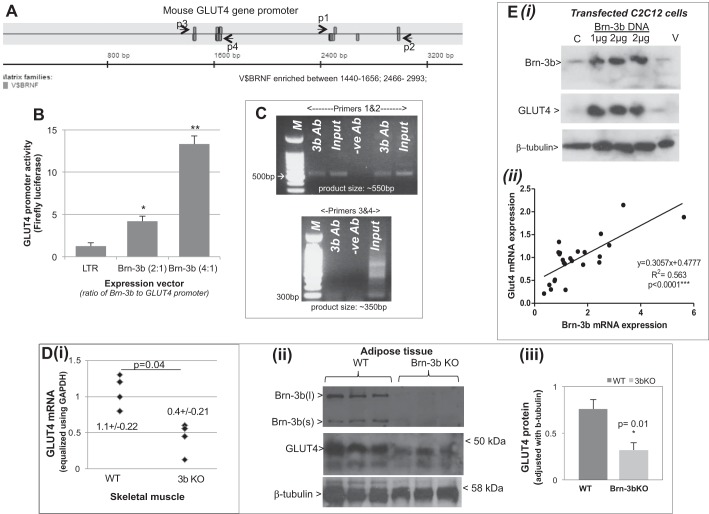
Fig. 5.
Brn-3b can activate GLUT4 expression. A: schematic diagram to show positions of potential Brn-3b binding sites (V$BRNF) in the mouse GLUT4 promoter sequence as determined by Genomatix Transfac analysis for transcription factor binding sites. Primers were designed to span each of 2 clusters of binding sites, with p1 and p2 flanking the proximal sites while p3 and p4 flanked the sites at the distal end of the promoter. B: results of luciferase assays showing effect of cotransfecting different ratios of Brn-3b expression vector with GLUT4 promoter reporter construct (2:1 or 4:1) into C2C12 cells. Values were equalized with internal control, Renilla luciferase activity and expressed relative to the promoter activity with empty expression vector (LTR; set at 1). Data shown represent means ± SD from at least 3 independent experiments. C: PCR products obtained when using ChIP DNA immunoprecipitated with Brn-3b or GAPDH antibody (negative control). Primers 1 and 2 amplified ∼500-bp PCR product from the proximal promoter, whereas primers 3 and 4 were designed to amplify the distal region of the promoter. D(i): results of qRT-PCR showing changes in GLUT4 mRNA levels in skeletal muscle prepared from Brn-3b KO mutant mice (n = 5) compared with WT littermate controls (n = 4). Variability in total RNA from different tissues was equalized using GAPDH. (ii): representative immunoblots showing changes in protein expression in Brn-3b KO adipose tissue compared with WT controls. Quantification of GLUT4 protein expression in Brn-3b KO tissue and age-matched WT control was adjusted with invariant protein β-tubulin. *Statistical significance P < 0.01 (t-test analysis). E(i): representative immunoblot showing GLUT4 protein levels in untransfected control C2C12 (C) in cells transfected with 1 or 2 μg of Brn-3b expression with empty expression or empty vector (V). (ii): regression analysis showing relationship between GLUT4 and Brn-3b mRNA expression in C2C12 cells grown under different conditions (n > 6 independent points).