Figure 1. Overview of the study.
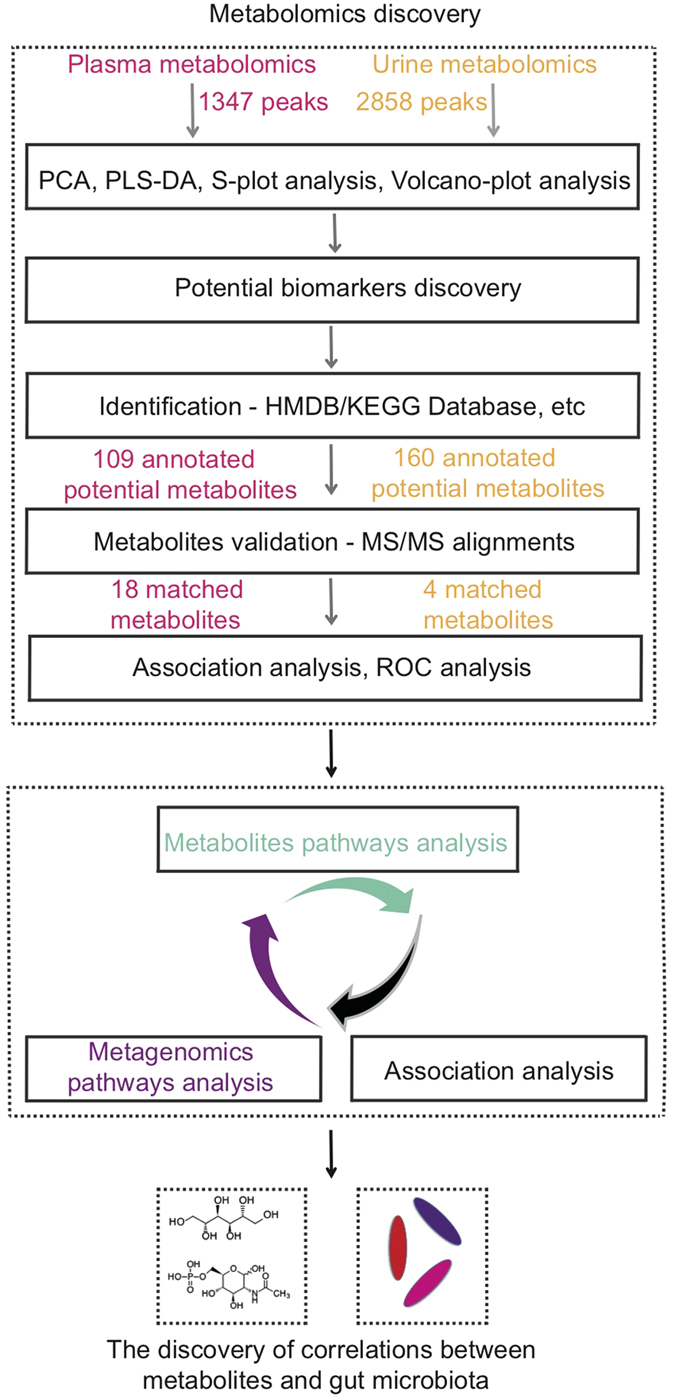
Non-targeted metabolomics technique is performed to discover potential metabolites in plasma and urine samples. Statistical and bioinformatics methods are used to identify significantly different metabolites that can discriminate CHD cases from healthy controls. Hierarchical cluster analysis (HCA) is performed to identify metabolites clusters contributing to phenotype separation and spearman correlation analysis is applied to identify potential biomarkers’ correlations related to abnormal functions. Pathway analysis and association analysis of potential biomarkers and gut flora are then applied. Finally, potential biomarkers associated gut flora species are discovered. Metagenomics technology is applied to further validate the potential metabolites originated from the fecal metagenomics data of CHD patients and healthy subjects.
