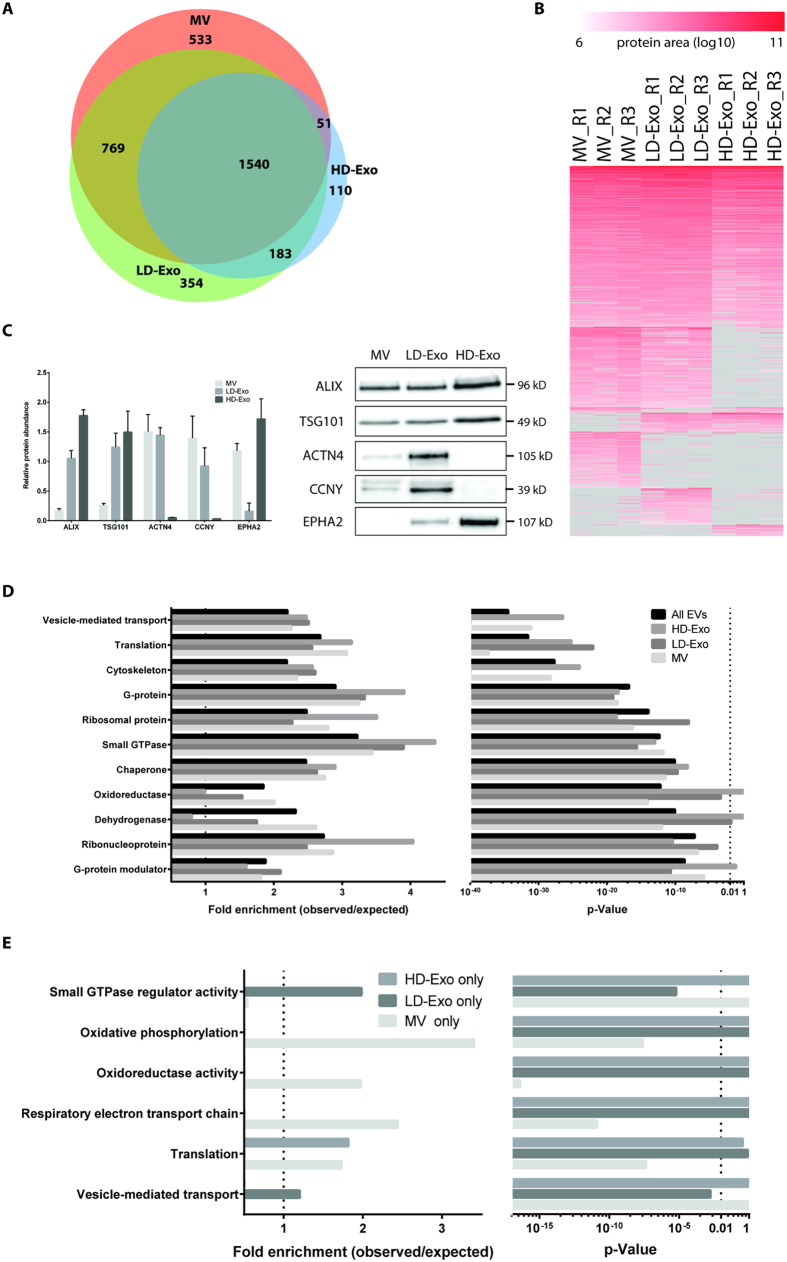
Figure 3. Proteome characterization of EV subpopulations by nanoLC-MS/MS reveals differences in protein composition.
(A) Overlap of identifications between MV, LD-Exo and HD-Exo. Proteins were considered identified if they had quantitative protein area in at least 2 of the 3 replicates. (B) Heatmap showing protein intensity (area) for each replicate. Subpopulations were grouped based on overlap in identifications as in (A), and subsequently ranked from highest to lowest average protein area. (C) Validation of proteomics data for selected proteins (left panel) by Western blotting (right panel). Equal amounts of protein were analyzed. (D) GO enrichments of all identified proteins and proteins identified in MV, LD-Exo and HD-Exo. (E) GO enrichments of proteins uniquely identified in MV, LD-Exo and HD-Exo.