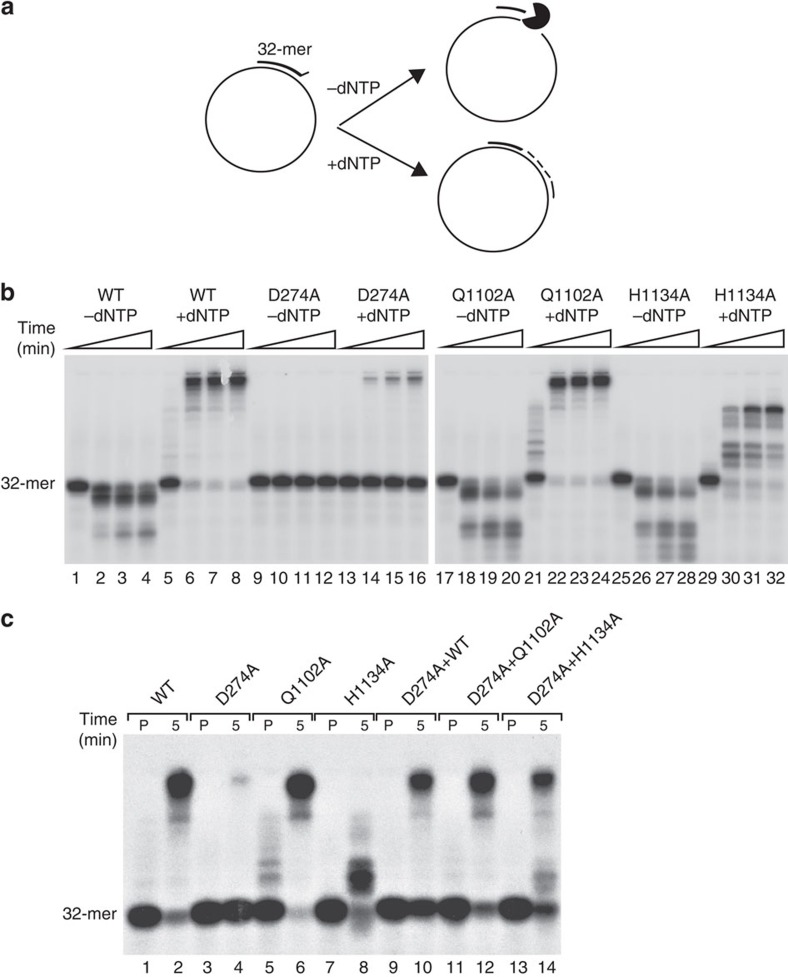
Figure 8. Mutant recombinant HsPOLγA proteins can complement each other in in vitro assays.
(a) Schematic overview of the HsPOLγA in vitro proofreading and polymerase activity assays. The template used in these assays consisted of a 5′ 32P-labelled 32-mer annealed to single-stranded pBluescript SK(+) creating one-nucleotide mismatch at the 3′-end. In the absence of dNTPs, HsPOLγA will backtrack (3′ to 5′ direction), degrading the oligonucleotide whereas in the presence of dNTPs the polymerase will proofread the mismatch and polymerize from the 3′-end. (b) The proofreading-deficient D274A had no exonuclease activity (lane 9–12), which led to impaired ability to continue DNA synthesis from the mismatched primer end (lane 13–16). The Q1102A mutant showed both extensive proofreading (lane 17–20) and polymerase activities (lane 21–24) comparable to the WT enzyme (lane 1–8). The H1134A mutant had extensive exonuclease activity (lane 25–28) but was a poor polymerase only able to copy a short stretch of DNA (lane 29–32). Triangles indicate elapsed time with 15 min being the end point. (c) The D274A and H1134A mutants were not able to copy DNA alone from the mismatched primer end (lanes 4 and 8, respectively). However, by mixing the two mutant proteins we observed a synergistic effect (lane 14) suggesting cooperation at the mismatched primer-end. The cooperation of D274A with WT and Q1102A is not as pronounced as in this assay (lane 10 and 12). ‘P' means the samples were pre-incubated on ice for 5 min and then stopped. Reactions labelled ‘5' were also pre-incubated on ice for 5 min but followed by a 5-min incubation at 37 °C before stopping the reaction. All experiments were repeated at least three time.