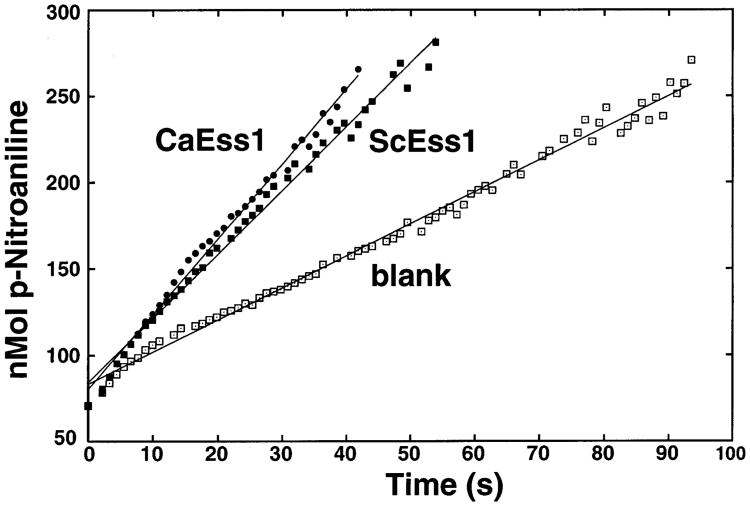
Figure 1.
PPIase activity of C. albicans Ess1 using a standard protease-coupled assay. The substrate (Suc-AEPF-pNA) was subjected to proteolysis by chymotrypsin, which releases the p-nitroaniline (pNA) from the peptide only when the E–P bond is in the trans conformation. The release of pNA was monitored during the assay at 20 °C by its absorbance at 390 nm, and the data were linearized as described in Experimental Procedures. Shown are representative graphs of experiments carried out in triplicate. The lines indicate the least-squares fit of the data; only every 15th data point is shown for clarity. The blank indicates no isomerase enzyme added. The results indicate that CaEss1 has activity comparable to that of the control S. cerevisiae enzyme (ScEss1).