Figure 6.
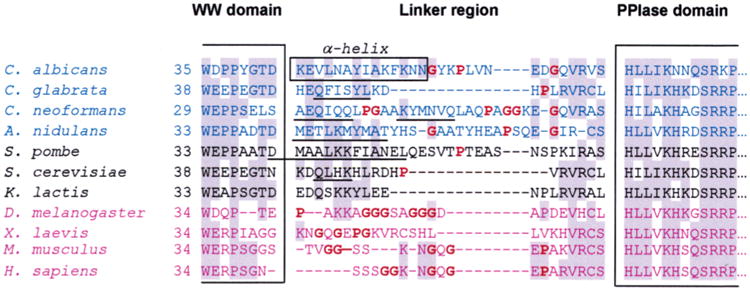
Sequence alignment of the linker region of Ess1 and Pin1 orthologs from different organisms. The sequences were aligned on the basis of highly conserved motifs within the WW and PPIase domains, some of which are shown within the large brackets. Shaded residues are the most highly conserved. The box denotes the residues in C. albicans that form a long α-helix in the crystal structure. Sequences in blue are from pathogenic fungi, and those in black are from other fungi that are not normally pathogenic. Sequences in purple are from metazoans. Bold residues (also highlighted in red) are helix-breaking residues or residues that do not promote helices (proline or glycine). Secondary structure predictions were performed for all linker regions using NNPREDICT (58). Sequences that are predicted to be α-helical are underlined. In general, fungal linker regions are predicted to include α-helices, whereas metazoan linkers probably do not.
