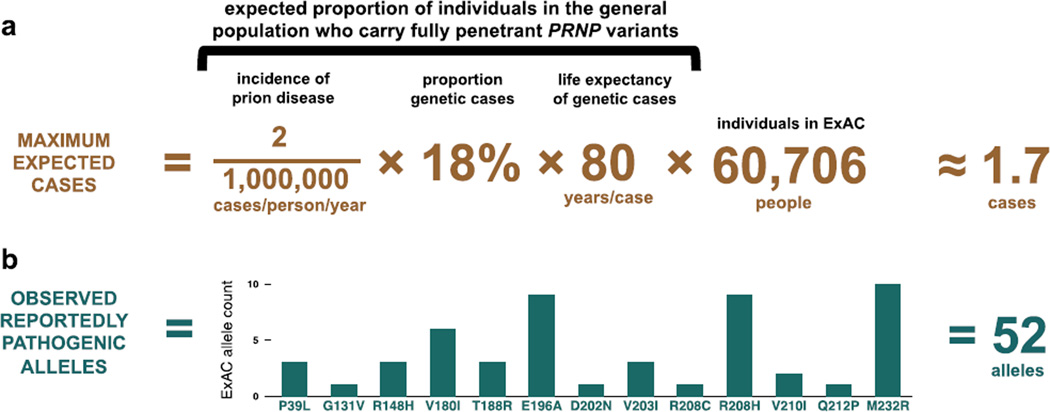
Figure 1. Reportedly pathogenic PRNP variants are >30 times more common in controls than expected based on disease incidence.
Reported prion disease incidence varies with the intensity of surveillance efforts (13), with an apparent upper bound of ~2 cases per million population per year (Materials and Methods). In our surveillance cohorts, 65% of cases underwent PRNP open reading frame sequencing, with 12% of all cases, or 18% of sequenced cases, possessing a rare variant (Table S1), consistent with an oft-cited estimate that 15% of cases of Creutzfeldt-Jakob disease are familial (31). Genetic prion diseases typically strike in midlife, with mean age of onset for different variants ranging from 28 to 77 (22, 32) (Table S10); we accepted 80, a typical human life expectancy, as an upper bound for mean age of onset, and to be additionally conservative, we assumed that all individuals in ExAC and 23andMe were below any age of onset, even though both contain elderly individuals (33) (Figure S1). Thus, no more than ~29 people per million in the general population should harbor high-penetrance prion disease-causing variants. Therefore at most ~1.7 people in ExAC (A) and ~15 people in 23andMe would be expected to harbor such variants. In fact, reportedly pathogenic variants are seen in 52 ExAC individuals (B) and on 141 alleles in the 23andMe database.