Abstract
Enzymatic “machines,” such as catalytic rods or colloids, can self-propel and interact by generating gradients of their substrates. We theoretically investigate the behaviors of such machines in a chemically active environment where their catalytic substrates are continuously synthesized and destroyed, as occurs in living cells. We show how the kinetic properties of the medium modulate self-propulsion and pairwise interactions between machines, with the latter controlled by a tunable characteristic interaction range analogous to the Debye screening length in an electrolytic solution. Finally, we discuss the effective force arising between interacting machines and possible biological applications, such as partitioning of bacterial plasmids.
I. INTRODUCTION
The interior of a living cell is a nonequilibrium medium that continuously produces and destroys high-energy molecules. These high-energy “solute” molecules may be bound and processed by enzymatic (biocatalytic) “machines.” Gradients of the solute molecules may a ect the self-organization of these catalytic cellular machines. Intriguingly, the biochemical activity of the underlying medium could alter such gradients, and thus modulate the behavior of these machines.
Self-propulsion and self-organization of catalytic colloidal particles through chemical gradients have previously been explored in synthetic systems without ambient chemical activity. Patterned catalytic particles can self-propel as artificial micro- or nano-motors by generating concentration gradients through chemical reactions at their surfaces [1-9]. Catalytic particles can also self-organize into a variety of nonequilibrium phases by interacting with the chemical gradients generated by other catalytic particles [10-13].
Additionally, the dynamics of protein catalysts (enzymes) can be altered in vitro by the presence of their substrates. Experiments have shown that the diffusion of enzyme molecules is enhanced when enzymes catalyze their substrates and that these molecular machines translocate up substrate gradients toward high substrate concentration [14-17]. These phenomena may have phoretic origins, although the precise mechanisms are not yet fully understood [14-18].
Phoretic effects of this general type are thought to play a role in organizing cellular structures and processes in vivo [19-24]. The cellular interior is a nonequilibrium environment with ambient chemical activity or passive biomolecule degradation, but previous studies have generally only considered an “inert” medium. In an inert environment, the medium itself neither creates nor destroys the substrate required for the machines’ catalytic behavior. However, molecular dynamics simulations have shown that the chemical activity of the underlying environment may alter diffusiophoresis [25, 26]. Those simulations illustrate that background chemical activity can reduce the self-propulsion speed [25]. Here, we analyze how the chemical activity of the environment controls self-propulsion and interactions of catalytically active machines.
In Section II we introduce a simple first-order kinetic model of catalysis by the environment, which immediately leads to a characteristic length scale, λ, set by the balance of solute diffusion and destruction, analogous to the Debye screening length in electrostatics. We investigate phoretic self-propulsion of the machines and find that asymmetric machines self-propel with a velocity that scales in a way that is dependent on the ratio of machine size to λ, as well as other factors. In Section III we analyze the pairwise interactions between machines. The magnitude and range of these interactions is regulated by the screening length found in Section II, and interactions can be tuned to be repulsive or attractive and strong or weak by chemical properties of the medium itself. Additionally, we construct a simulation of discrete solute particles and catalytic machines to demonstrate the validity of our continuous-field approach, and show that an effective potential can be used to describe machine interactions in the appropriate physical regime. In Section IV we discuss the application of our model to a biological subcellular spatial organization problem of current interest, namely the partitioning of plasmid DNA molecules within a bacterium.
II. SELF-PROPULSION
A. Theoretical model
We consider diffusiophoresis of enzymatic particles in a chemically active medium which generates reactant solute at rate k+ and destroys it at rate of k−c, where c is the local reactant concentration. The system is overdamped; the solute species diffuses with constant Ds, and spherical catalytic particles of radius r0 diffuse slowly with constant Dc ≪ Ds. The catalytic particles interact with the solute species chemically by creating or destroying solute and physically by attracting or repelling solute. The model predicts that bulk biochemical activity alters self-propulsion and controls the range, strength, and sign of interactions between catalytic particles.
We first analyze the model in the continuum limit for the case of a single catalytic particle. As in the analysis of the inert medium (where k+ = k− = 0) in [3], we determine the solute concentration near the catalytic particle. Since Dc ≪ Ds, we assume a steady-state concentration profile. Thus, we solve a reaction-diffusion equation:
| (1) |
which accounts for both diffusion and the chemical activity of the medium.
At the surface of the catalytic particle, we consider two distinct scenarios. In the first, solute concentration is sufficiently high so that the surface catalysis rate is reaction-limited, and thus independent of the local solute concentration [3]. We also consider a diffusion-limited case where the surface catalysis rate grows linearly with local solute concentration; this case only has a nontrivial steady-state in a chemically active medium. These cases correspond to two different boundary conditions:
| (2) |
where the catalytic activity of the particle is given by the surface flux, α(θ), or the surface flux per unit solute concentration, γ(θ). These quantities are positive or negative for, respectively, solute creation or destruction, and may vary with the polar angle θ on the particle (we assume azimuthal symmetry). We assume c|∞ = k+/k− is the bulk steady state far from the catalytic particle.
Solving Eq. (1) in three dimensions (see Appendix A for results in dimensions d = 1 and d = 2), we find the series:
| (3) |
where kℓ is the ℓth modified spherical Bessel function of the second kind [27, 28], Pℓ is the ℓth Legendre polynomial, bℓ is a coefficient, and the decay length is the typical distance that solute diffuses before being destroyed.
The perturbation of the solute concentration field due to the catalytic particle decays as
| (4) |
On short length scales, the concentration field is similar to that of the inert medium [3], but on length scales longer than the decay length, the concentration field decays exponentially due to the activity of the medium.
We determine the coefficients, {bℓ}, from Eq. (2) by expanding the surface activity in Legendre polynomials [3]. In the reaction-limited case,
| (5) |
where αℓ is the ℓth coefficient in the expansion of α(θ). In the diffusion-limited case, bℓ is determined by solving the system of equations:
| (6) |
with given by [29]. The infinite set of equations in Eq. (6) can be solved in closed form to any desired precision by setting bℓ = 0 for ℓ larger than a cuto ℓ* in the expansion of γ [30].
When the catalytic activity of the particle is asymmetric, it can self-propel. To determine the self-propulsion speed we calculate the slip velocity, vs(θ) [3]. vs is the difference between the particle velocity and the velocity of the ambient fluid at the surface of the particle [31]; this slip is responsible for net particle propulsion [3, 31, 32]. vs arises from the tangential component of the concentration gradient at the particle surface [3, 31]:
| (7) |
where μ(θ) is the mobility coefficient [3, 31].
Using a version of the low Reynolds number reciprocal theorem [3, 32, 33], we find that the propulsion velocity, , is the slip velocity averaged over the surface of the particle (see Appendix B):
| (8) |
Substituting for the slip velocity, , and integrating, we have:
| (9) |
where {μℓ} are the coefficients of the Legendre polynomials in the expansion of μ(θ).
B. Reaction-limited case
For the reaction-limited case, there are two scaling regimes for the propulsion speed, v:
| (10) |
The speed is proportional to the catalytic activity of the particle and the strength of the physical interaction between the particle and the solute species.
In the limit of slow bulk solute degradation, the activity of the medium is negligible over the length of the catalytic particle, which is small compared to solute decay length. Thus, v scales as in the inert medium, where self-propulsion is independent of particle size [3] because the concentration profile is scale-free on the scale of r0.
When the catalytic particle is larger than the decay length, medium chemical activity qualitatively alters particle self-propulsion by perturbing the solute concentration. Thus, v decreases as particle size, r0, and/or the bulk solute degradation rate, k−, increase. Unlike the size-dependence that arises with Michaelis-Menten surface kinetics [34], the r0-dependence here arises from the importance of the activity of the underlying medium over length scales > λ. Eq. (10) demonstrates that the chemical properties of the medium, in addition to its physical properties such as viscosity, η ∝ 1/Ds, control self-propulsion of catalytic particles.
C. Diffusion-limited case
For diffusion-limited systems, self-propulsion depends on the chemical properties of the medium for all particle sizes. For the simplest case with self-propulsion, we consider the minimal asymmetric model with γ(θ) = γ0 + γ1P1(cos θ).
To simply describe the system, we reduce the parameters by nondimensionalizing the reaction-diffusion equation by rescaling lengths by and concentrations by c0 = k+/k−:
| (11) |
Thus, the boundary condition, Eq. (2b), is written as:
| (12) |
where R = r0/λ is the dimensionless particle size, and and are the dimensionless catalysis rates.
The concentration is now written as:
| (13) |
where . Consequently, the self-propulsion speed scales as:
| (14) |
where the coefficient is found through a systematic approximate solution to the system of equations, Eq. (6) (see Appendix C).
For sufficiently small g0 and g1, defined approximately as , the self-propulsion speed scales as (see Appendix C):
| (15) |
v is insensitive to γ0 at leading order, but it varies linearly in γ0 for small γ0 and as for very negative γ0 (inset to Fig. 1a). Thus, for slow surface catalysis, v scales as in the reaction-limited case, Eq. (10), but multiplied by the solute concentration, c0 = k+/k−, sustained by the medium.
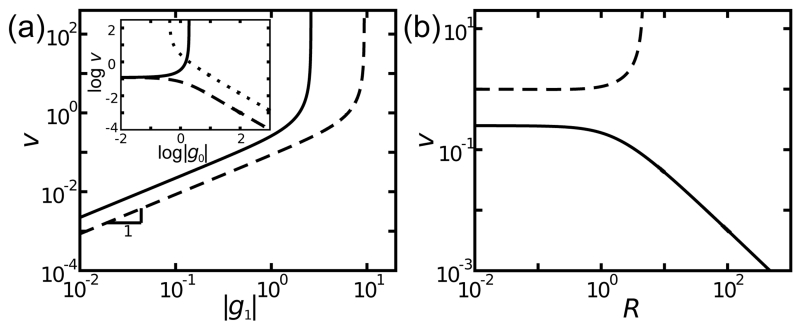
FIG. 1. Scaling of v for diffusion-limited cases.
a) v (in units of μc0/r0) grows linearly in |g1| until diverging. Solid and dashed lines show v with g0 = ±|g0|, respectively. Inset: v for g0 > 0 with small |g1| (solid), g0 < 0 with small |g1| (dashed), and g0 < 0 with large |g1| (dotted). v grows linearly with |g0| for small |g0| if g1 is sufficiently small, but decays as 1/|g0| for sufficiently negative g0 (dashed) or diverges for large |g0| (solid). For large |g1|, steady-state v exists only for sufficiently negative g0 (dotted). b) v is insensitive to R at small R, but decays at large R as 1/R if g0 and |g1| are sufficiently small (solid) and diverges otherwise (dashed).
Rapid solute production by the catalytic particle can overwhelm the destruction of solute by the medium. In that case, the model assumptions are not compatible with a nonequilibrium steady-state. As in Fig. 1a, v diverges for large g0 and |g1|. Similarly, for large g0 and |g1|, v diverges for large particles since the total surface catalysis overwhelms background activity (Fig. 1b). Mathematically, these divergences arise from the zeros of the denominator of the expression for the coefficient (see Eq. (C1)). In a real autocatalytic system (e.g., actin filament networks [35]), surface catalysis saturates at high concentration, and thus, the system should transition to a steady-state with the boundary condition in Eq. (2a) and velocity in Eq. (10).
III. INTERACTIONS
A. Screening of interactions
Catalytic particles are capable of self-organization through their interactions with the concentration fields generated by other catalytic particles [10-13, 36]. In an inert medium, these interactions are long-ranged, decaying as 1/r (Eq. (4a)) [12, 13]. In a chemically active medium, however, the solute distribution decays exponentially (Eq. (4b)). A similar screening length arises in the limit of many closely-packed catalytic particles in the diffusion-limited (“unsaturated”) regime, which is discussed in [13]. Thus, as with electrostatic interactions in an ionic solution, diffusiophoretic interactions in an active medium are exponentially screened.
B. Simulation model
To understand interactions in a biochemically active medium, we developed a Brownian dynamics simulation model in which catalytic particles chemically and physically interact with rapidly diffusing solute molecules. We consider catalytic particles of radius r0 that repel each other by excluded volume interactions in a periodic system of size L. These particles act as potential wells for diffusing solute molecules, binding solute within a distance r0 with energy ε. While bound, solute molecules are degraded by the catalytic particle at rate . Solute particles diffuse with coefficient Ds = 1000Dc, are degraded by the medium at rate k− when not bound to a catalytic particle, and are produced by the medium at rate k+. The coupling of physical and chemical interactions models a collection of enzymes, each of which must bind its substrate before catalyzing a reaction. Additional model details are provided in Appendix D. Although some details of the simulation differ from the theory, the simulation can be described by the theory with an appropriate choice of surface activity, as described in Appendix E.
The interactions between two diffusiophoretic particles depend on the whether the particles locally deplete or enhance the solute concentration and whether they attract or repel solute [12]. In the simulation, the effective surface activity, α, is proportional to the ambient concentration k+/k− and the composite rate, , comparing solute degradation by the medium to solute degradation and sequestration (binding) by the catalytic particle (Eqs. (E9) and (E10)). Thus, enzyme-like machines in a biochemically active medium repel each other when , attract when , and do not interact when .
A catalytic particle locally depletes solute by binding it with energy ε and degrading it at rate . The concentration of solute bound to the catalytic particle is times larger than the local medium solute concentration; this enhances the rate of solute depletion by the particle by . Thus, even if , solute can be depleted from the medium if it only rarely unbinds from the catalytic particle (large ε). However, if catalytic particles degrade solute more slowly than the medium and the binding time is short (small ε), the particle behaves as solute source. This demonstrates that interactions, and thus self-organization, may be altered through manipulation of either the active medium or the catalytic particle properties.
C. Simulation results and theoretical analysis
To quantify interactions, we construct an effective equilibrium theory incorporating both excluded volume and concentration-gradient-mediated interactions. For simplicity, we consider the simplest case with symmetric particles, which do not independently self-propel (i.e., only considering the ℓ = 0 terms of the Legendre polynomial expansions of α(θ) and μ(θ)). However, the analysis can be generalized to specific cases of asymmetric, self-propulsive particles as well (by considering the ℓ > 0 terms in the expansions).
Excluded volume directly induces repulsive forces on length scales r < 2r0, which leads to an effective entropic repulsion over longer length scales. Catalytic particles also interact through their concentration fields. Thus, the effective interaction potential between particles is:
| (16) |
where c(r) is the single-particle concentration profile (e.g., Eq. (E1) for d = 3 or Eq. (E11) for d = 1) and w is the volume of the solute-binding region of the machine. Since the solute concentration is continuously in steady state with respect to the catalytic particle positions, it acts as a static potential. The coefficient, ε, is the binding energy.
For , this approach is justified by the separation of timescales between particle motions and concentration profile perturbations. In particular, the effective equilibrium approximation used to estimate interactions between catalytic particles relies on the separation of the characteristic timescale, τc, for catalytic particle motion from the lifetime, τs, of perturbations to the solute concentration profile.
The timescale for catalytic particle motion is given by the diffusion coefficient as: . Since the particles are symmetric, they do not self-propel, so there is no need to consider an effective temperature description [37] based on the intrinsic velocity of the catalytic particles.
The lifetime for perturbations to the solute concentration profile can be calculated by considering linear perturbations to the reaction-diffusion equation (Eq. (1)):
| (17) |
Writing , we find , implying .
For the timescales to be comparable, we must have . This is only satisfied for real q if ; otherwise for all real q, as desired for the equilibrium approximation. Since Ds ≫ Dc by assumption, τc ≈ τs(q) requires either the catalytic particle to be very small or the bulk solute decay rate to be very slow. Thus, the assumption of separation of timescales is valid if . This is satisfied in our simulations, where and λ/r0 ≈ 2.2 (moreover, the slowest τs(q) is τs(q = 0) = 20, which is much shorter than τc = 4000).
As a further consistency check, we consider the timescale for particle motion due to the effects of the concentration fields generated by other particles. This time scale is , where the speed, |v|, is calculated as the magnitude of the effective force, , divided by the particle drag coefficient, kBT/Dc. Since and w = 2r0 in one dimension, we have .
From Eq. (E12), which gives the effective surface catalysis rate in the simulation, we note that |α| < k+ max(r0, λ). Since Ds/Dc ≫ 1, we also have unless ε, k+, r0 or λ is very large. For the simulations considered, we have compared to τs(q) ≤ 20.
To directly test the equilibrium approximation, we compare the theory with the simulation in the case where the two machines are pushed together by an external force, F . In the theory, the inter-particle force-distance relation is given by the statistical-mechanical relation:
| (18) |
which can be expressed exactly for the inert medium (see Appendix F).
Distance fluctuations, , can be derived in a similar manner (Eq. (F4)). As shown for the one-dimensional examples in Fig. 2, the theory (lines) agrees with the simulation (circles) provided that the concentration near each particle is large enough that the continuum approximation is valid and small number fluctuations are unimportant. This requires , similar to Debye-Huckel theory.
FIG. 2. Force-distance relation and distance fluctuations from theory and simulation.
a) Without chemical activity, particles pushed together with force F (units of 2kBT/r0) entropically repel (solid line). 〈r〉 as a function of F is depressed for attractions in both the theory (dashed line) and simulation (solid circles). 〈r〉 is larger for repulsions in the theory (dash-dotted line) and simulation (open circles). Dotted line shows 〈r〉 = 2r0 and L = 30. b) The corresponding distance fluctuations.
Using the equilibrium theory, we studied how changes in the underlying medium affect the mean distance, 〈r〉, and fluctuations, 〈δr2〉, between interacting catalytic particles in d = 1 (Fig. 3). 〈r〉 varies non-monotonically with k− (Fig. 3a, solid line). For slow bulk degradation (), particles are repulsive, so 〈r〉 is larger than the zero-activity mean distance, L/4 + r0 (dotted line). With fast bulk degradation (), particles are attractive, so 〈r〉 decreases to 2r0. For very fast k−, the solute concentration is low, so particle interactions vanish and 〈r〉 approaches its non-interacting value.
FIG. 3. Medium activity controls mean distance and fluctuations.
a) 〈r〉 varies non-monotonically with k− (solid line). 〈r〉 ≈ L/2 for strong repulsions and 〈r〉 ≈ 2r0 for strong attractions. For weak interactions, i.e., (vertical line) or large k−, 〈r〉 → L/4 + 2r0 (dotted). b) is small for and , but large for and large k−. c) As k+ increases, 〈r〉 decreases/increases toward maximum compression/extension for attractions/repulsions (solid/dashed). d) For large k+, decreases as 1/k+ for attractions (solid) and for repulsions (dashed). L = 50 in all panels.
Inter-particle distance fluctuations also vary non-monotonically with k− (Fig. 3b, solid line). For k− such that the attractions or repulsions are strong, fluctuations are small, since the particles are tightly held in their equilibrium positions. However, for , the interactions are weak, so 〈δr2〉 approaches the inert limit, 〈δr2〉 = (L – 4r0)2/48. The particles also interact weakly and exhibit large distance fluctuations for large k−, when the ambient concentration is low.
As the bulk production rate, k+, increases, interaction strength grows linearly, and particles transition from weakly to strongly interacting. However, changes to k+ cannot change the sign of interactions. Thus, 〈r〉 is nearly the inert particle limit for small k+, but for large k+, approaches maximum compression or separation for attractive or repulsive interactions, respectively (Fig. 3c).
The increase in interaction strength with increasing k+ manifests as a power law decay in fluctuations (Fig. 3d). For large for attractive interactions and for repulsive interactions. The power laws differ because attractive interactions are dominated by the shape of the solute concentration field at short length scales, whereas repulsive interactions are dominated by its shape at long length scales.
D. Effective force
To determine whether diffusiophoretic interactions are relevant in biological systems, we estimate the force arising from the d = 3 solute concentration profile (again assuming separation of timescales). The effective force due to diffusiophoretic interactions can be estimated by considering the solute concentration profile generated by the combined activity of the catalytic particles and the underlying medium.
In d = 3, the concentration profile surrounding a catalytic particle with surface activity α (in the reaction-limited system) or γ (in the diffusion-limited system) is given by Eq. (E1). In the reaction-limited case (see Appendix G for the diffusion-limited case), the resulting radial concentration gradient is:
| (19) |
We estimate the characteristic effective force, F*, due to concentration-gradient mediated interactions by εw∂rc at one screening length from the surface of the catalytic particle (r = r0 + λ):
| (20) |
Estimating the volume of interaction between particle and solute as (as in the simulation model), we have:
| (21) |
Thus, we find that F* scales as:
| (22) |
Assuming ε ≈ few kBT , Ds ≈ 1 μm2/s, k− = 0.1 s−1, and α = 0.01 nm−2 s−1 (i.e., 1 enzyme with catalysis rate 1 s−1 per 100 nm2), F * ≈ 1 pN for μm-sized particles. We therefore conclude that diffusiophoretic interactions can be important in biological systems.
IV. APPLICATION TO BACTERIAL PLASMID PARTITIONING
Experiments and models suggest that chromosome segregation, plasmid partitioning, and positioning of organelle-like structures by the bacterial ParAB system are phoretically driven by a ParA protein gradient [21, 22, 24, 38-43]. In plasmid partitioning, the protein ParB binds to parS-sites of plasmid DNA while ParA binds the nucleoid. ParA molecules diffuse along the nucleoid until dissociating into the cytoplasm. The ParB-parS complex binds ParA, stimulating faster ParA dissociation from the nucleoid. This system spaces plasmids equally throughout the rod-shaped bacterium [44].
We interpret this as phoretic interactions between plasmids. Since ParB sequesters and dissociates ParA from the surrounding nucleoid, it locally depletes ParA, as observed in vivo [45] and in vitro [24, 40, 41]. This induces effective repulsions between ParB-bound plasmids, which move toward high ParA concentration. ParA polymerization and depolymerization [46], may quantitatively, but not qualitatively, alter this mechanism.
The bacterium is approximately a one-dimensional system. For such a system, the effective force, F* = εw∂xc, may be derived from the one-dimensional concentration profile, approximately given by L → ∞ limit of Eq. (E11). Thus, we estimate the effective force with w = r0, as:
| (23) |
For the ParAB plasmid partitioning system, where the catalytic particle binds and degrades solute as described for the simulation, the effective surface catalysis rate is approximately given by the L → ∞ limit of Eq. (E12):
| (24) |
To estimate the interaction range and effective force, we use measurements from in vitro and in vivo experiments. We assume Ds = 0.85 μm2/s and k− = 0.032 s−1, which were measured in vitro [41]. To estimate the association rate, which is the effective production rate, we estimate 2400 ParA molecules per nucleoid as measured in [43] and estimate that the nucleoid is 3 μm in length. We set this (d = 1) concentration equal to k+/k− in order to arrive at k+ = 25 s−1μm−1, which is consistent with the estimate in [43] and also the in vitro experiments in [38, 40, 41]. We estimate that ε ≈ few kBT , consistent with the dynamics of ParA-ParB-parS complexes observed in in vitro experiments [40, 41, 47]. We estimate kc− ≈ 10k− which is approximately the rate at which ParB-stimulated hydrolysis of ParA-ATP occurs in vitro [47, 48]; however, we note that using a higher rate, kc−, as in [43] increases α by an factor. Finally, we estimate r0 ≈ 0.1 μm based on in vivo fluorescence images such as those in [43].
We thus estimate an interaction range of λ ≈ 5 μm and, by Eq. (23), we estimate an effective force of order 10 fN. λ is long enough to facilitate plasmid interactions. F * is comparable to the ≈ 10 fN viscous drag that plasmids experience in vivo [49], and sufficient to self-organize plasmids against thermal fluctuations.
V. CONCLUSION
Our model demonstrates a novel mechanism for controlling self-propulsion and self-organization. The chemical activity of the underlying medium affects these behaviors by altering the steady-state solute concentration field. Tuning the background activity can change both the strength and the sign of interparticle interactions, and define length scales of order λ, which may be much larger than the size of the solute molecules or the enzymatic machines. For the ParAB bacterial plasmid partitioning system, our estimates suggest that the relevant length scale, λ, is in the range of microns, and forces on protein-coated plasmids are in the ten femtonewton range. In general, the effective force generated by catalytic machines in other in vivo scenarios could be as large as a few piconewtons.
Our results suggest a number of general comments. First, we anticipate that catalytic particle properties (transport, interactions, and self-organization) within cells might be controlled by altering the composition of macromolecular complexes or post-translational protein modifications (PTPMs, e.g., phosphorylation). Second, since the behavior of the catalytic machines is dependent on their environment and its activity as well, we expect transport and interaction effects from changes in cytoplasmic properties. Such “medium” properties might be manipulated by (over)expressing enzymes that degrade reactant “solute” molecules. In terms of intracellular regulation, cells could change bulk concentrations and kinetics of small biomolecules such as NTPs or proteins with specific NTPs and PTPMs to modulate organization of catalytic particles or organelles. Finally, we note that at supracellular scales, chemotactic cells such as bacteria or immune cells may utilize similar principles to those discussed in this paper to mediate swarming or dispersive interactions.
ACKNOWLEDGMENTS
We acknowledge support by NSF Grants MCB-1022117 and DMR-1206868, and by NIH Grants 1U54CA193419 and 1R01GM105847. This research was supported in part through the computational resources and staff contributions provided for the Quest high performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Appendix A: Reaction-diffusion equation in d = 1 and d = 2
In d = 1, the solution to Eq. (1)) is:
| (A1) |
where + and − subscripts, respectively, denote the solutions for x > r0 and x < −r0.
In d = 2, we have:
| (A2) |
where Kℓ is a modified Bessel function of the second kind.
Appendix B: Derivation of the self-propulsion velocity
As described in [32], the reciprocal theorem for low Reynolds number hydrodynamics [33] may be used to solve for the velocity, , of self-propelled particle in terms of the drag force, acting against motion and the fluid velocity, , at the surface of the object:
| (B1) |
where σ is the stress on the particle surface due to motion against the fluid and the integration is performed over the surface of the particle.
In the model, the surface velocity, , is the slip velocity, , arising from the tangential component of the concentration gradient at the particle surface [3]. Thus, for a spherical particle with and , we find that the propulsion velocity is the slip velocity averaged over the surface of the particle:
| (B2) |
Substituting for the slip velocity, , we have:
| (B3) |
where we have expanded μ(θ) in Legendre polynomials. Integrating this expression leads to Eq. (9).
Appendix C: Scaling of the self-propulsion velocity in the diffusion-limited system
In order to approximately solve Eq. (6) in a systematic manner, we assume that bℓ≥2 = 0. Thus, the coefficient is approximately:
| (C1) |
where R, g0, and g1 are reduced variables as described in Section II C.
We first consider the limit of reduced catalysis rates, g0 and g1, that are sufficiently small so that the catalysis of the particle does not overwhelm the catalysis of the medium. Thus, we only have physical solutions when g0 is sufficiently small or negative and |g1| is sufficiently small. The maximum values of these parameters is given by the smallest real root of the denominator in Eq. (C1) (i.e., the values of g0 and g1 for which diverges).
There is a maximum value for g0; on the other hand, if g0 is very negative, the catalytic particle destroys solute very quickly, which leads to a valid steady-state with low solute concentration. If g0 were too large, concentration-dependent solute production by the catalytic particle could overcome the concentration-dependent destruction of solute in the medium.
For g1, there is an upper limit to its absolute value; since P1(cos θ) = cos θ is positive for one half of the particle surface, but negative for the other half, g1 is related to both destruction and production of solute, regardless of its sign. Moreover, the expression for is manifestly odd in g1. Since the total rates of solute catalysis at the particle surface by the ℓ = 1 term depend on the local solute concentration, if |g1| is too large, production of solute on one side of the catalytic particle can overwhelm solute destruction on the other side of the particle and the entire medium. In that case, there is no physically-meaningful steady-state solution to the reaction-diffusion equation, and the solute concentration grows without bound. We note that in a real system, this unphysical behavior would be cut off by physical effects not considered in this formulation of the boundary condition, such as saturation of the activity at the particle surface.
The denominator in Eq. (C1) is zero for
| (C2) |
and
| (C3) |
As discussed above, g0 must be small enough or sufficiently negative to overcome the solute production due to g1, so g0 must be less than the “–” expression in Eq. (C2). In contrast, since |g1| must be small, we require that g1 lie between the “+” and “–” solutions of Eq. (C3).
To simplify these expressions, we first combine the small and large R limits of Eq. (C2) to obtain an approximate threshold. This yields , which requires g0 and |g1| strictly less than the actual thresholds in Eqs. (C2)-(C3).
Since is unphysical (and moreover, Eq. (C1) is approximate, with the approximation worsening near the divergence in ), we assume that g0 and g1 are small enough that we are far from these threshold values.
With these assumptions, for R ≪ 1, we find:
| (C4) |
which is the exact expression for to . (The exact expression for to may be found by calculating under the assumption b3 = 0, etc.)
Thus, for provided that (so that the does not dominate) and (so that does not dominate if g0 ≈ 0).
For R ≫ 1, we have:
| (C5) |
which scales as for small g0 and |g1|.
By combining these scaling expressions with the scaling of k1(R) with R, we arrive at Eq. (15).
Finally, the scaling with g0 can be extracted by noting that near , whereas for .
Appendix D: Simulation details
The catalytic particles of radius r0 effiectively maintain excluded volume interactions through a short-ranged harmonic repulsion, which is given by:
| (D1) |
for r < 2r0, where K = 500kBT.
Catalytic particles bind solute particles through a Lennard-Jones-like potential, which is truncated at r = rc so that energy and force are zero at the edge of the potential well:
| (D2) |
where fixes the minimum to a. The minimum energy is approximately −ε. We set a = 1 and rc = r0 = 2 in our simulations.
All particles in the simulation are also subject to Brownian noise, F (t), which is described by:
| (D3) |
| (D4) |
where ζ is the particle drag.
The equations of motion are given by [50]:
| (D5) |
which we integrate using an Euler algorithm.
Appendix E: Mapping theory onto simulation
In the theoretical model described above, symmetric catalytic particles produce the following steady-state concentration fields:
| (E1) |
The steady-state solute concentration distribution in the simulation corresponds to the following set of reaction-diffusion equations:
| (E2) |
| (E3) |
The matching conditions at the boundary, r = r0, are approximately given by:
| (E4) |
| (E5) |
Note that this is an approximation because the potential given in Eq. (D2) is smooth rather than infinitely sharp at r = r0.
In a system of infinite size in d = 3, the solution to these equations is:
| (E6) |
where and i0 is the 0th modified spherical Bessel function of the first kind (), and B0 and b0 are constants determined from the matching conditions. These constants are:
| (E7) |
| (E8) |
This solution (Eqs. (E6)-(E8)) can be rewritten in the form of Eq. (E1). To do this, choose
| (E9) |
for the solution to the reaction-limited system, or equivalently, choose
| (E10) |
for the diffusion-limited system. It can be shown that denominator is not zero for any real positive Ds, k−, , ε, and r0 so this mapping is well-behaved.
In a periodic system of size L in d = 1, corresponding to the simulation examples given in the text, we have the additional constraints that the concentration is periodic in and so that the concentration varies smoothly everywhere. Considering a catalytic particle centered on r = 0 and denoting the effective surface catalysis rate (in the reaction-limited scenario) as α, we have exponential solutions for the concentration profile:
| (E11) |
where the effective surface catalysis rate is:
| (E12) |
Eqs. (E11)-(E12) completely describe the steady-state solute concentration profile in d = 1 as a function of the simulation parameters k+, k−, , Ds, L, r0, and ε.
Appendix F: Calculation of the force-distance and force-fluctuation relations
Here we consider the mean distance, 〈r〉, between two thermally-diffusing, chemically-inert, hard particles of radius r0 pushed together by force F in a periodic system of length L. The mean distance diverges as F −1 as the force pushing the particles together approaches 0. According to equilibrium statistical mechanics, 〈r〉 can be found by integrating:
| (F1) |
Thus, we find:
| (F2) |
where the brackets contain the value of 〈r〉 in the L → ∞ limit. In general dimension, we can concisely write:
| (F3) |
where is the incomplete gamma function [27].
Similarly, fluctuations can be calculated as
| (F4) |
which diverges as 1/(βF)2 in the L → ∞ system.
Appendix G: Effective force in the diffusion-limited case
In d = 3, the concentration profile surrounding a catalytic particle with surface activity α (in the reaction-limited system) or γ (in the diffusion-limited system) is given by Eqs. (E1). In the reaction-limited case (see Appendix (G for the diffusion-limited case), the resulting radial concentration gradient is:
| (G1) |
Thus, using Eq. (20), we find the characteristic force in the diffusion-limited case to be:
| (G2) |
This force scales as:
| (G3) |
References
- [1].Paxton WF, Kistler KC, Olmeda CC, Sen A, St. Angelo SK, Cao Y, Mallouk TE, Lammert PE, Crespi VH. J. Am. Chem. Soc. 2004;126:13424. doi: 10.1021/ja047697z. [DOI] [PubMed] [Google Scholar]
- [2].Golestanian R, Liverpool TB, Ajdari A. Phys. Rev. Lett. 2005;94:220801. doi: 10.1103/PhysRevLett.94.220801. [DOI] [PubMed] [Google Scholar]
- [3].Golestanian R, Liverpool TB, Ajdari A. New J. Phys. 2007;9:126. [Google Scholar]
- [4].Howse JR, Jones RAL, Ryan AJ, Gough T, Vafabakhsh R, Golestanian R. Phys. Rev. Lett. 2007;99:048102. doi: 10.1103/PhysRevLett.99.048102. [DOI] [PubMed] [Google Scholar]
- [5].Córdova-Figueroa UM, Brady JF. Phys. Rev. Lett. 2008;100:158303. doi: 10.1103/PhysRevLett.100.158303. [DOI] [PubMed] [Google Scholar]
- [6].Jülicher F, Prost J. Eur. Phys. J. E. 2009;29:27. doi: 10.1140/epje/i2008-10446-8. [DOI] [PubMed] [Google Scholar]
- [7].Popescu MN, Dietrich S, Tasinkevych M, Ralston J. Eur. Phys. J. E. 2010;31:351. doi: 10.1140/epje/i2010-10593-3. [DOI] [PubMed] [Google Scholar]
- [8].Sabass B, Seifert U. Phys. Rev. Lett. 2010;105:218103. doi: 10.1103/PhysRevLett.105.218103. [DOI] [PubMed] [Google Scholar]
- [9].Michelin S, Lauga E. J. Fluid Mech. 2014;747:572. [Google Scholar]
- [10].Theurkau I, Cottin-Bizonne C, Palacci J, Ybert C, Bocquet L. Phys. Rev. Lett. 2012;108:268303. doi: 10.1103/PhysRevLett.108.268303. [DOI] [PubMed] [Google Scholar]
- [11].Palacci J, Sacanna S, Steinberg AP, Pine DJ, Chaikin PM. Science. 2013;339:936. doi: 10.1126/science.1230020. [DOI] [PubMed] [Google Scholar]
- [12].Soto R, Golestanian R. Phys. Rev. Lett. 2014;112:068301. doi: 10.1103/PhysRevLett.112.068301. [DOI] [PubMed] [Google Scholar]
- [13].Saha S, Golestanian R, Ramaswamy S. Phys. Rev. E. 2014;89:062316. doi: 10.1103/PhysRevE.89.062316. [DOI] [PubMed] [Google Scholar]
- [14].Yu H, Jo K, Kounovsky KL, de Pablo JJ, Schwartz DC. J. Am. Chem. Soc. 2009;131:5722. doi: 10.1021/ja900372m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Muddana HS, Sengupta S, Mallouk TE, Sen A, Butler PJ. J. Am. Chem. Soc. 2010;132:2110. doi: 10.1021/ja908773a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sengupta S, Dey KK, Muddana HS, Tabouillot T, Ibele ME, Butler PJ, Sen A. J. Am. Chem. Soc. 2013;135:1406. doi: 10.1021/ja3091615. [DOI] [PubMed] [Google Scholar]
- [17].Riedel C, Gabizon R, Wilson CAM, Hamadani K, Tsekouras K, Marqusee S, S. P, Bustamante C. Nature. 2015;517:227. doi: 10.1038/nature14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Golestanian R. Phys. Rev. Lett. 2015;115:108102. doi: 10.1103/PhysRevLett.115.108102. [DOI] [PubMed] [Google Scholar]
- [19].Lee KC, Liu AJ. Biophys. J. 2008;95:4529. doi: 10.1529/biophysj.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]; 2009;97:1295. [Google Scholar]
- [20].Alexander GP, Liu AJ. “Self-diffusiophoresis in the advection dominated regime,”. 2011 arXiv:1107.3851. [Google Scholar]
- [21].Banigan EJ, Gelbart MA, Gitai Z, Wingreen NS, Liu AJ. PLoS Comput. Biol. 2011;7:e1002145. doi: 10.1371/journal.pcbi.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sugawara T, Kaneko K. Biophysics. 2011;7:77. doi: 10.2142/biophysics.7.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Banigan EJ, Lee KC, Liu AJ. Phys. Biol. 2013;10:066004. doi: 10.1088/1478-3975/10/6/066004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vecchiarelli AG, Neuman KC, Mizuuchi K. Proc. Natl. Acad. Sci. USA. 2014;111:4880. doi: 10.1073/pnas.1401025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thakur S, Kapral R. J. Chem. Phys. 2011;135:024509. doi: 10.1063/1.3607408. [DOI] [PubMed] [Google Scholar]
- [26].Robertson B, Kapral R. J. Chem. Phys. 2015;142:154902. doi: 10.1063/1.4918329. [DOI] [PubMed] [Google Scholar]
- [27].Arfken GB, J H. Weber, Mathematical Methods for Physicists. Elsevier Academic Press; New York: 2005. [Google Scholar]
- [28].Modified spherical Bessel functions of the second kind may be written as [27].
- [29].Adams JC. Proc. R. Soc. Lond. 1878;27:63. the product of the nth and mth Legendre polynomials can be expanded in Legendre polynomials. The coefficient of the ℓth Legendre polynomial is: , where and when s is integral and the sum of any two of n, m, and ℓ is not greater than the third; otherwise cnmℓ = 0.
- [30].The values of bℓ fall off rapidly. For example, for in the large ℓ limit, and typically decay at least exponentially for ℓ > 1. In general, bℓ is small for ℓ > ℓmax, where ℓmax denotes the highest-order term in the expansion of γ(θ).
- [31].Anderson JL. Annu. Rev. Fluid Mech. 1989;21:61. [Google Scholar]
- [32].Stone HA, Samuel ADT. Phys. Rev. Lett. 1996;77:4102. doi: 10.1103/PhysRevLett.77.4102. [DOI] [PubMed] [Google Scholar]
- [33].Happel J, Brenner H. Low Reynolds Number Hydrodynamics. Martinus Nijho Publishers; The Hague: 1983. [Google Scholar]
- [34].Ebbens S, Tu MH, Howse JR, Golestanian R. Phys. Rev. E. 2012;85:020401(R). doi: 10.1103/PhysRevE.85.020401. [DOI] [PubMed] [Google Scholar]
- [35].Carlsson AE. Biophys. J. 2003;84:2907. doi: 10.1016/S0006-3495(03)70018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phys. Rev. Lett. 2004;92:238102. doi: 10.1103/PhysRevLett.92.238102. [DOI] [PubMed] [Google Scholar]
- [36].Pohl O, Stark H. Phys. Rev. Lett. 2014;112:238303. doi: 10.1103/PhysRevLett.112.238303. [DOI] [PubMed] [Google Scholar]
- [37].Tailleur J, Cates ME. Phys. Rev. Lett. 2008;100:218103. doi: 10.1103/PhysRevLett.100.218103. [DOI] [PubMed] [Google Scholar]; Europhys. Lett. 2009;86:60002. [Google Scholar]
- [38].Vecchiarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, Biertümpfel C, Funnell BE, Mizuuchi K. Mol. Microbiol. 2010;78:78. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vecchiarelli AG, Mizuuchi K, Funnell BE. Mol. Microbiol. 2012;86:513. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hwang LC, Vecchiarelli AG, Han YW, Mizuuchi M, Harada Y, Funnell BE, Mizuuchi K. EMBO J. 2013;32:1238. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vecchiarelli AG, Hwang LC, Mizuuchi K. Proc. Natl. Acad. Sci. USA. 2013;110:E1390. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kiekebusch D, Thanbichler M. Trends Microbiol. 2014;22:65. doi: 10.1016/j.tim.2013.11.005. [DOI] [PubMed] [Google Scholar]
- [43].Ietswaart R, Szardenings F, Gerdes K, Howard M. PLoS Comput. Biol. 2014;10:e1004009. doi: 10.1371/journal.pcbi.1004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ebersbach G, Gerdes K. Mol. Microbiol. 2004;52:385. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- [45].Hatano T, Yoshiharu Y, Niki H. Mol. Microbiol. 2007;64:1198. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ringgaard S, van Zon J, Howard M, Gerdes K. Proc. Natl. Acad. Sci. USA. 2009;106:19369. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Havey JC, Vecchiarelli AG, Funnell BE. Nucleic Acids Res. 2012;40:801. doi: 10.1093/nar/gkr747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Easter J, Jr., Gober JW. Mol. Cell. 2002;10:427. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- [49].To estimate the viscous drag force felt by a plasmid, we note that plasmids move at speeds of approximately v = 1 μm/min (e.g., see [45]), and have a drag coefficient of approximately kBT/Dplasmid, where Dplasmaid> ≈ 10−3 μm2/s [43]. This leads to an estimated drag force of order 10 fN.
- [50].Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; New York: 1989. [Google Scholar]