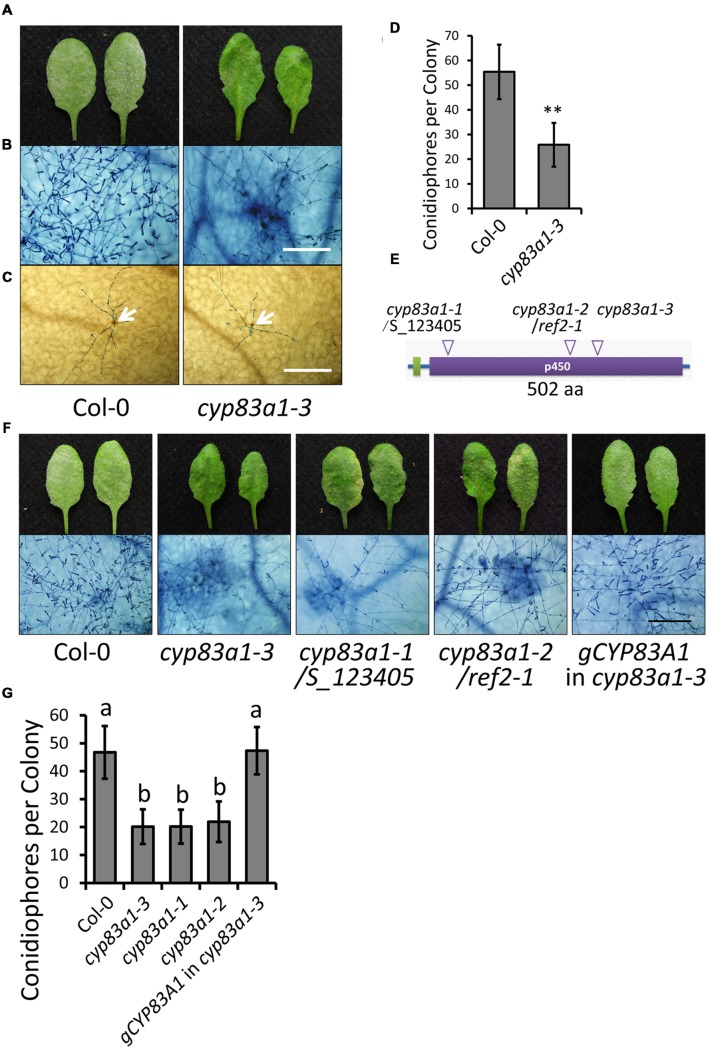
FIGURE 1.
The cyp83a1-3 mutant displays enhanced resistance to Golovinomyces cichoracearum. (A) Four-weeks-old Arabidopsis wild-type and cyp83a1-3 mutant plants were infected with G. cichoracearum, and representative leaves were removed and photographed at 8 dpi. (B) Leaves were stained with trypan blue at 8 dpi, bar = 200 μm. (C) Leaves were stained with DAB and trypan blue at 2 dpi; arrows indicate H2O2 accumulation, bar = 200 μm. (D) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independents experiments (n = 30). Asterisk represents statistically significant differences from wild-type (P < 0.01, nested ANOVA). (E) Schematic representation of the CYP83A1 protein, arrows indicate mutation sites of three cyp83a1 mutant alleles. (F) Four-weeks-old wild-type, cyp83a1-3, cyp83a1-1, cyp83a1-2, and transgenic cyp83a1-3 mutant plants complemented with the wild-type CYP83A1 gene (gCYP83A1) were infected with G. cichoracearum and representative leaves were stained with trypan blue at 8 dpi, bar = 200 μm. (G) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Results represent the mean and standard deviation in three independent experiments (n = 30; P < 0.01, nested ANOVA).