Abstract
MicroRNAs are important in cancer development and progression. In the present study, the clinical significance and function of microRNA-711 (miR-711) expression in breast cancer were investigated. The expression level of miR-711 was analyzed in breast cancer tissue samples using reverse transcription-quantitative polymerase chain reaction. Cell proliferation, colony formation, apoptosis and Transwell assays were performed in breast cancer cell lines transfected with miR-711 mimics or inhibitors, or control sequence. miR-711 was found to be upregulated in 30 formalin-fixed paraffin-embedded breast cancer tissue samples compared with paired non-cancerous breast tissues (P<0.05). Furthermore, a higher miR-711 expression was demonstrated to be associated with poor overall and disease-free survival times in 161 breast cancer patients, and miR-711 was identified as an independent prognostic factor using multivariate Cox regression analysis. In vitro, overexpression of miR-711 resulted in a significant increase in proliferation, colony formation, migration and invasion of breast cancer cells. By contrast, downregulating miR-711 inhibited cell proliferation, colony formation, migration and invasion and enhanced the rate of apoptosis of breast cancer cells. To the best of our knowledge, the present study is the first to demonstrate that miR-711 is an independent prognostic factor and serves an important oncogenic function in breast cancer, suggesting that miR-711 is a potential biomarker of prognosis and a molecular therapeutic target in breast cancer.
Keywords: breast cancer, miR-711, prognosis, cell proliferation, reverse transcription-quantitative PCR
Introduction
Breast cancer is the most common cancer and the leading cause of cancer-associated mortality in women worldwide (1). Breast cancer mortality rates have decreased in North America and several European countries over the past 25 years, principally due to earlier detection and improved treatments (2,3). However, in numerous African and Asian countries, the incidence and mortality rates have increased (4). Therefore, mortality from breast cancer remains a significant health issue globally. In general, the primary risk factors for breast cancer in women are old age (>50 years) and circulating estrogen (5). There are two strategies for enhancing the survival of breast cancer patients: Early detection and appropriate treatment. Primary treatments include surgery, radiotherapy, chemotherapy, endocrine therapy and targeted therapy. Studies have shown that the integrated use of a variety of treatments is advantageous to the prognosis of breast cancer patients (6). At present, formulation of a treatment plan, including surgery and adjuvant therapy, is primarily based on clinical stage and the presence of several limited biomarkers, including estrogen receptor, progesterone receptor and human epidermal growth factor receptor-2 (6). However, the tumor-node-metastasis staging system and these biomarkers do not meet the requirements for personalized therapy and/or precise therapy. Identifying biomarkers for precisely predicting prognosis or for future targeted therapy is one of the major challenges in this disease.
Previous studies have demonstrated that a large proportion of human microRNAs (miRNAs) are located in cancer-associated regions of the genome (7), which implies that miRNAs are important in the pathogenesis of various human cancers (8–14). miRNA expression is usually dysregulated in various cancer types (15,16), including breast cancer (17). miRNAs specifically bind to the 3′-untranslated region of target mRNAs, which results in the degradation of the mRNA or inhibition of its translation (18). In addition, miRNAs may be capable of affecting the transcription of target genes through promoter activation (19) or transcriptional silencing (20). Therefore, miRNAs are critical in cancer diagnosis, prognosis and individualized therapy (21).
In the present study, miR-711 expression was detected using reverse transcription (RT)-quantitative polymerase chain reaction (qPCR) in 30 pairs of formalin-fixed paraffin-embedded (FFPE) breast cancer and non-cancerous matched tissue samples. miR-711 is located on chromosome 3. The majority of previous research concerning miR-711 has focused on organ injury (22–24), including myocardial infarction. The findings indicated that miR-711 was overexpressed in breast cancer. Therefore, the present study revealed a functional role of miR-711 in human cancer, particularly in breast cancer. In addition, analysis of 161 FFPE breast cancer samples demonstrated that increased levels of miR-711 expression were associated with poor overall survival time of patients. Overall, the present in vitro findings suggest that miR-711 has an oncogenic role in breast cancer cells.
Materials and methods
Patients and tissue samples
For the current study, 30 paired FFPE breast cancer and non-cancerous samples were obtained from patients who underwent surgical treatment for breast cancer between January 2004 and December 2005 at Sun Yat-sen University Cancer Center (Guangzhou, Guangdong, China). In addition, 161 FFPE breast cancer samples were obtained from patients who underwent radical mastectomy between June 2002 and December 2006 at the First Affiliated Hospital of Guangzhou Medical University (Guangzhou, Guangdong, China). All breast cancer samples were pathologically diagnosed as invasive ductal carcinoma by two experienced pathologists. None of the 161 patients had distant metastasis at diagnosis. The percentage of tumor cells in the FFPE breast cancer tissues was >70%. The 161 patients had not received radiotherapy or chemotherapy prior to mastectomy. Clinical staging was based on the American Joint Committee on Cancer staging manual (25). The clinical characteristics of the 161 patients are summarized in Table I. The median follow-up time for patients from the First Affiliated Hospital of Guangzhou Medical University was 75.2 months (range, 6.7–115.8). Overall survival (OS) time was calculated between the date of surgery and the date of mortality due to any cause or of last follow-up, and the disease-free survival (DFS) time was calculated between the date of surgery and the date of the first distant metastasis, relapse or mortality due to any cause. The study was reviewed and approved by the Ethics Committees of Sun Yat-sen University Cancer Center and the First Affiliated Hospital of Guangzhou Medical University.
Table I.
Association between miR-711 expression level and clinical characteristics.
| miR-711 expressiona | |||
|---|---|---|---|
| Clinical characteristics | High, n (%) | Low, n (%) | P-value |
| All patients | 80 (100) | 81 (100) | |
| Age, years | 0.88 | ||
| >45 | 43 (54) | 45 (56) | |
| ≤45 | 37 (46) | 36 (44) | |
| Menopause | 0.82 | ||
| No | 42 (52) | 44 (54) | |
| Yes | 38 (48) | 37 (46) | |
| Pathological grade | 0.52 | ||
| I | 2 (3) | 5 (6) | |
| II | 68 (85) | 66 (82) | |
| III | 10 (13) | 10 (12) | |
| Estrogen receptor | 0.21 | ||
| Positive | 55 (69) | 48 (59) | |
| Negative | 25 (31) | 33 (41) | |
| Progesterone receptor | 0.58 | ||
| Positive | 48 (60) | 52 (64) | |
| Negative | 32 (40) | 29 (36) | |
| HER2 | 0.69 | ||
| Positive | 30 (37) | 28 (34) | |
| Negative | 50 (63) | 53 (66) | |
| Tumor stage | 0.23 | ||
| T1 | 17 (21) | 19 (24) | |
| T2 | 47 (59) | 55 (68) | |
| T3 | 14 (18) | 6 (7) | |
| T4 | 2 (3) | 1 (1) | |
| Node stage | 0.10 | ||
| N0 | 36 (45) | 47 (58) | |
| N1 | 23 (29) | 25 (31) | |
| N2 | 15 (19) | 6 (7) | |
| N3 | 6 (8) | 3 (4) | |
| TNM stage | 0.17 | ||
| I | 11 (14) | 15 (19) | |
| II | 43 (54) | 50 (62) | |
| III | 26 (33) | 16 (20) | |
| Radiotherapy | 0.35 | ||
| No | 16 (20) | 22 (27) | |
| Yes | 64 (80) | 59 (73) | |
| Chemotherapy | 0.86 | ||
| No | 24 (30) | 23 (28) | |
| Yes | 56 (70) | 58 (72) | |
χ2 test or Fisher's exact test used for P-value calculation.
Patients were divided into a high or low miR-711 expression group using the median expression level as the cut-off point (0.57). miR-711, microRNA-711; HER2, human epidermal growth factor receptor 2; TNM; tumor-node-metastasis.
Total RNA extraction
The breast cancer and non-cancerous tissues were cut into 8–10 µm sections, deparaffinized and rehydrated. Total RNA was extracted from the tissue sections using a modified phenol/chloroform extraction method, as previously described (26). Total RNA from cell lines was extracted using Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. A NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used to estimate the concentration and quality of total RNA.
RT-qPCR
Reverse transcription was performed on the 30 paired tissue samples using 10 ng of total RNA, 1X miRNA-specific reverse transcription primers (Thermo Fisher Scientific, Inc.), 100 µM nucleoside triphosphates (Thermo Fisher Scientific, Inc.), 3.33 U/µl MultiScribe™ Reverse Transcriptase (Thermo Fisher Scientific, Inc.), 1X Reverse Transcription Buffer (Thermo Fisher Scientific, Inc.) and 1.33 U/µl RNase inhibitor (Thermo Fisher Scientific, Inc.) in a final volume of 15 µl. The reaction was conducted at 16°C for 30 min, followed by 30 min at 42°C and 5 min at 85°C. The qPCR reaction was performed using a 20 µl volume containing 1.33 µl reverse transcription products, 1X TaqMan® Small RNA Assay solution (including specific primers and probes; Applied Biosystems; Thermo Fisher Scientific, Inc.) and 1X Universal PCR Master Mix II (no UNG; Thermo Fisher Scientific, Inc.). The RT-qPCR was performed in triplicate for each sample using an Applied Biosystems PRISM 7900HT System (Thermo Fisher Scientific, Inc.) with the following conditions: 50°C for 2 min; 95°C for 10 min; and 45 cycles of 95°C for 15 sec and 60°C for 60 sec. The primer sequences used were as follows: MiR-711 forward, 5′-ACACTCCAGCTGGGGGGACCCAGGGAGAGA-3′; and reverse, 5′-TGGTGTCGTGGAGTCG-3′. Small nuclear RNA U6 was used as a normalization control. The 2−ΔΔCq equation was used to represent the relative expression of miRNA (27) and Student's t-test was used to analyze the results.
Cell culture and transfection
MCF-7, MDA-MB-231 and ZR-75-30 human breast cancer cell lines were cultured in Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% Gibco fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C with an atmosphere of 5% CO2. The miR-711 mimic, inhibitor and miRNA negative control (NC) were designed and synthesized by Shanghai GenePharma, Ltd. (Shanghai, China). When the cells reached 60–70% confluence, Invitrogen Lipofectamine® 2000 RNAiMAX reagent (Thermo Fisher Scientific, Inc.) was used to perform the transfection of cells with 100 nM miR-711 mimic or inhibitor, or NC, according to the manufacturer's protocol.
Cell growth
Following transfection for 36 h, the cells were cultured in 96-well plates (1,000 cells/well) for an additional 7 days. The growth of the cells was measured using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. At a specific time every day for 7 days, 20 µl MTT (dilution, 0.5 mg/ml; Weijia Biology Science and Technology Co., Ltd, Guangzhou, China) was added, followed by additional incubation for 4 h at 37°C. Following the removal of the supernatant, 150 µl dimethyl sulfoxide (Weijia Biology Science and Technology Co., Ltd) was added to each well. The cells in the 96-well plate were gently agitated for 10 min and the absorbance was measured at 490 nm (SpectraMax® M5 Multi-Mode Microplate Reader; Molecular Devices LLC, Sunnyvale, CA, USA) to calculate the cell growth rate.
Colony formation assay
At 36 h subsequent to transfection, MCF-7 cells were seeded on a 6-well plate (1,000 cells/well) and incubated at 37°C in an atmosphere of 5% CO2 for 2 weeks. The surviving colonies (>50 cells/colony) were counted using crystal violet staining.
Apoptosis assay
Following transfection with miR-711 inhibitors for 48 h, the cells were washed twice with ice-cold phosphate-buffered saline and resuspended in 400 µl 1X Binding Buffer (Bestbio Co., Shanghai, China). The cells were stained with Annexin V-fluorescein isothiocyanate (Bestbio Co.) for 15 min at 4°C. Subsequently, propidium iodide was added to the cells for 5 min at 4°C. The percentage of apoptotic cells was analyzed using flow cytometry (FC 500; Beckman Coulter, Inc., Brea, CA, USA).
In vitro migration and invasion assays
A Boyden chamber Transwell assay (24-well Transwell plate; BD Biosciences, Franklin Lakes, NJ, USA) was used to measure cell migration and invasion, and was performed according to the manufacturer's protocol. For Transwell migration, 5×104 MDA-MB-231 cells were plated into the top chamber of the Transwell plate in DMEM without FBS. The bottom chamber of the Transwell plate was filled with DMEM supplemented with 10% FBS to stimulate migration. Following an incubation period of 16 h, the cells in the membrane of the bottom chamber were fixed with 100% methanol, stained with 0.1% crystal violet (Weijia Biology Science and Technology Co., Ltd) and counted under a microscope (BX53; Olympus Corp., Tokyo, Japan). For Transwell invasion, the membrane of the top chamber of the Transwell plate was coated with Matrigel (BD Biosciences) and 2×105 cells were plated into the top chamber. Following an incubation period of 24 h, the fixed and stained cells in the bottom chamber were counted.
Statistical analysis
The Kaplan-Meier method and the log rank test were used to analyze OS time and DFS using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Univariate and multivariate Cox regression analysis were performed to identify the independent prognostic factors for survival. The association between miR-711 and clinical characteristics of the patients was analyzed using Student's t-test, χ2 test or Fisher's exact test. SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for all analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-711 is upregulated in breast cancer
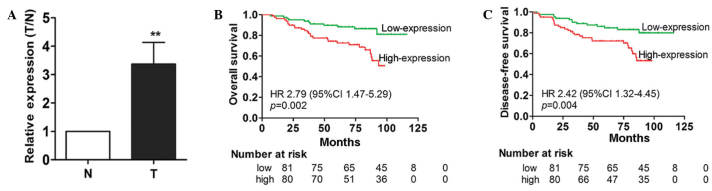
To investigate whether miR-711 is dysregulated in breast cancer, 30 paired FFPE breast cancer and non-cancer tissue samples were collected, and miR-711 expression was assessed in the samples using TaqMan RT-qPCR. The findings demonstrated that the expression level of miR-711 in breast cancer tissues was significantly increased compared with the paired non-cancerous breast tissues (Fig. 1A). This suggests that miR-711 may be a potential oncogene in breast cancer.
Figure 1.
miR-711 was upregulated in patients with breast cancer and was associated with a poor survival time. (A) Expression levels of miR-711 in 30 paired T and N tissues, detected using reverse transcription-quantitative polymerase chain reaction. The relative expression level was normalized to small nuclear RNA U6. Paired Student's t-test was performed; data are presented as the mean ± standard deviation; **P<0.01. (B) miR-711 expression in tissues from patients with breast cancer is associated with overall survival time. (C) miR-711 expression in tissues from patients with breast cancer is associated with disease-free survival of the patients. Log rank tests were performed to compare the survival times. miR-711, microRNA-711; T, breast cancer tissue; N, normal adjacent breast tissue.
miR-711 expression is associated with survival but not clinical characteristics of breast cancer patients
To additionally investigate the clinical significance of overexpressed miR-711, RT-qPCR analysis was conducted in 161 FFPE breast cancer samples. Based on the miR-711 expression levels measured by RT-qPCR, the 161 patients were divided into a high or low miR-711 expression group using the median expression level as the cut-off point (0.57; range, 0.23 to 1.21). The associations between miR-711 expression level and clinical characteristics were evaluated; however, no significant associations were observed (Table I). By contrast, a Kaplan-Meier survival analysis indicated that patients in the high miR-711 expression group demonstrated significantly poorer OS and DFS times compared with patients in the low-expression group (P=0.002 and P=0.004, respectively; Fig. 1B and C).
miR-711 is an independent prognostic factor for breast cancer
Based on the findings that miR-711 expression is significantly associated with survival time of patients with breast cancer, the present study investigated whether miR-711 is an independent prognostic factor for breast cancer. The effects of miR-711 expression and various clinical characteristics on the survival time of 161 patients was analyzed using univariate and multivariate Cox regression models. The results demonstrated that miR-711 expression was an independent prognostic factor for OS time [hazard ratio (HR), 2.549; 95% CI, 1.303–4.988; P=0.006] and DFS time (HR, 2.873; 95% CI, 1.392–5.929; P=0.004) in patients with breast cancer (Table II). Tumor-node-metastasis stage was also found to be an independent prognostic factor for OS (HR, 1.928; 95% CI, 1.162–3.198; P=0.011) and DFS (HR, 2.068; 95% CI, 1.209–3.536; P=0.008). Furthermore, miR-711 was not associated with other clinical characteristics, suggesting that this miRNA independently affects the survival time of patients with breast cancer. Overall, the results indicate that miR-711 is an independent prognostic factor in breast cancer.
Table II.
Univariate and multivariate Cox regression analysis for the identification of prognostic factors for OS and DFS time in patients with breast cancer.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Clinical feature | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Analysis for OS time | ||||
| Expression of miR-711 (high vs. low) | 1.985 (1.159–3.398) | 0.007 | 2.549 (1.303–4.988) | 0.006 |
| Age, years (>45 vs. ≤45) | 1.015 (0.944–1.137) | 0.654 | – | – |
| Menopause (yes vs. no) | 1.187 (0.555–2.539) | 0.659 | – | – |
| Pathological grade (III vs. I–II) | 1.426 (0.626–3.251) | 0.398 | – | – |
| HER2 (positive vs. negative) | 1.693 (1.082–2.260) | 0.080 | – | – |
| PR (positive vs. negative) | 1.045 (0.782–1.398) | 0.764 | – | – |
| ER (positive vs. negative) | 0.981 (0.737–1.307) | 0.897 | – | – |
| TNM stage (III vs. II vs. I) | 1.985 (1.159–3.398) | 0.012 | 1.928 (1.162–3.198) | 0.011 |
| Analysis for DFS time | ||||
| Expression of miR-711 (high vs. low) | 2.927 (1.406–6.092) | 0.004 | 2.873 (1.392–5.929) | 0.004 |
| Age, years (>45 vs. ≤45) | 1.021 (0.979–1.107) | 0.785 | – | – |
| Menopause (yes vs. no) | 1.377 (0.610–3.105) | 0.441 | – | – |
| Pathological grade (III vs. I–II) | 1.095 (0.421–2.848) | 0.852 | – | – |
| HER2 (positive vs. negative) | 1.310 (1.082–1.704) | 0.092 | – | – |
| PR (positive vs. negative) | 1.129 (0.826–1.543) | 0.448 | – | – |
| ER (positive vs. negative) | 0.923 (0.681–1.251) | 0.606 | – | – |
| TNM stage (III vs. II vs. I) | 2.235 (1.252–3.989) | 0.007 | 2.068 (1.209–3.536) | 0.008 |
n=161. miR-711, microRNA-711; HR, hazard ratio; CI, confidence interval; OS, overall survival; DFS, disease-free survival; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor; TNM; tumor-node-metastasis.
miR-711 overexpression promotes oncogenic growth of breast cancer cells
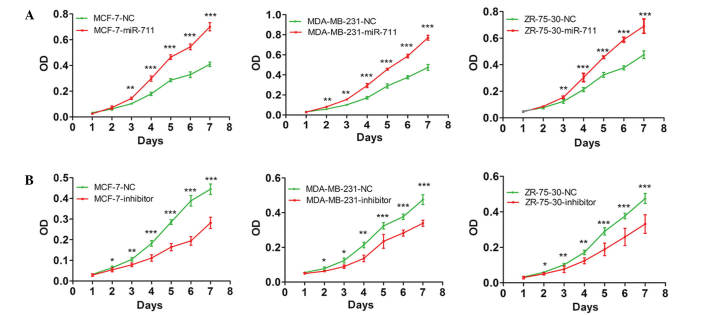
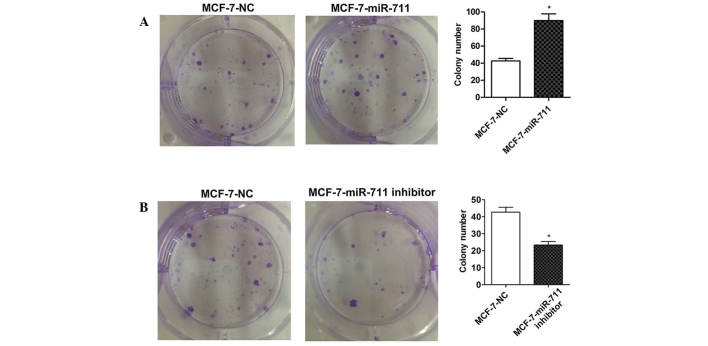
In order to understand how miR-711 may affect the survival time of breast cancer patients, the present study investigated the mechanisms by which miR-711 promotes the development and progression of breast cancer. MCF-7, MDA-MB-231 and ZR-75-30 human breast cancer cell lines were transiently transfected with a miR-711 mimic and the cell proliferation was observed. MTT assay demonstrated that the growth rate of the cells transfected with miR-711 mimic was significantly increased compared with cells transfected with the NC miRNA (P=0.002; Fig. 2A). Subsequently, cells transfected with a miR-711 inhibitor were analyzed. The findings demonstrated that the proliferation of the cells transfected with the miR-711 inhibitor were markedly decreased compared with cells transfected with NC (P=0.001; Fig. 2B). A colony formation assay was performed on MCF-7 cells transfected with a miR-711 mimic or inhibitor. As expected, the colony numbers of MCF-7 cells transiently transfected with miR-711 mimics was significantly increased compared with MCF-7 cells transfected with NC (P=0.007; Fig. 3A), and the colony numbers were significantly decreased in cells transfected with a miR-711 inhibitor compared with cells transfected with NC (P=0.008; Fig. 3B). These findings indicate that miR-711 promotes the oncogenic growth of breast cancer cells.
Figure 2.
miR-711 overexpression promotes the proliferation of breast cancer cells. (A) Human breast cancer MCF-7, MDA-MB-231 and ZR-75-30 cells were transiently transfected with a miR-711 mimic or NC and measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The growth curves demonstrated significantly different proliferation rates between the cells transfected with miR-711 and NC. (B) MCF-7, MDA-MB-231 and ZR-75-30 cells were transiently transfected with miR-711 inhibitor or NC. The growth curves demonstrated that downregulated miR-711 significantly reduced the proliferation rate of cells with the miR-711 inhibitor compared with cells transfected with NC. *P<0.05; **P<0.01; ***P<0.001. miR-711, microRNA-711 NC, microRNA negative control; OD, optical density.
Figure 3.
miR-711 overexpression enhances the colony formation of breast cancer cells. (A) Human breast cancer MCF-7 cells were transiently transfected with a miR-711 mimic or NC and a colony formation assay was used to determine the number of colonies. The number of colonies formed by the breast cancer cells transfected with miR-711 mimics markedly increased compared with the cells transfected with NC. (B) MCF-7 cells were transiently transfected with a miR-711 inhibitor or NC. The number of colonies formed by the breast cancer cells transfected with a miR-711 inhibitor was markedly reduced compared with the cells transfected with NC. *P<0.05. miR-711, microRNA-711; NC, negative control.
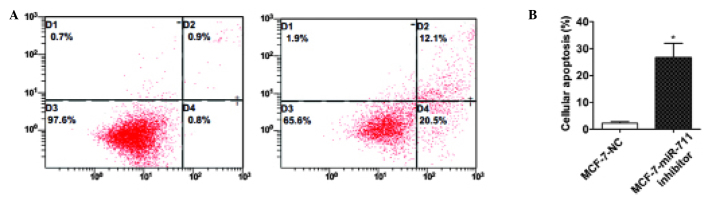
Knockdown of miR-711 induces apoptosis in breast cancer cells
The present study aimed to identify whether apoptosis affects the growth of breast cancer cells induced by miR-711. In the apoptosis assay, MCF-7 cells were transiently transfected with a miR-711 inhibitor or NC, and flow cytometry was used to evaluate the results. The findings demonstrated that the apoptotic rate of MCF-7 cells transfected with the miR-711 inhibitor was markedly increased compared with cells transfected with NC (Fig. 4). This suggests that downregulation of miR-711 may induce apoptosis in breast cancer cells. Therefore, miR-711 may inhibit cell apoptosis, leading to the proliferation of breast cancer cells.
Figure 4.
Downregulating miR-711 induces the apoptosis of breast cancer cells. (A) Human breast cancer MCF-7 cells were transiently transfected with a miR-711 inhibitor or NC and apoptotic cells were detected by flow cytometry. (B) The histogram indicates the rate of apoptosis in cells transfected with a miR-711 inhibitor or NC. miR-711, microRNA-711; NC, negative control.
miR-711 affects the migration and invasion of breast cancer cells
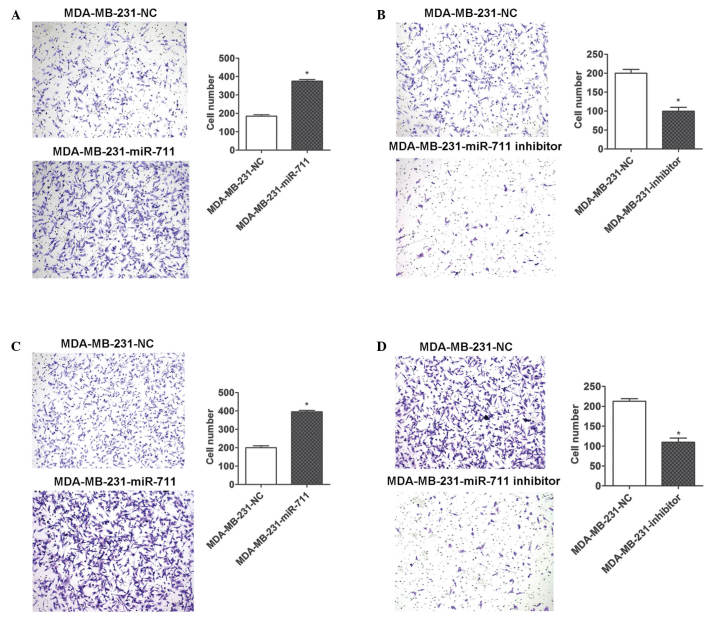
To additionally investigate miR-711 in the progression of breast cancer, migration and invasion assays were conducted using Transwell plates. MDA-MB-231 cells were transiently transfected with miR-711 mimic, inhibitor or NC and were seeded in the wells of the Transwell plate. The ectopic expression of miR-711 significantly promoted the migration and invasion of MDA-MB-231 cells (Fig. 5A and C), and the downregulation of miR-711 using a miR-711 inhibitor resulted in marked inhibition of migration and invasion of the cells (Fig. 5B and D). Therefore, the present results indicate that miR-711 serves a functional role in tumor progression and metastasis, which is consistent with the poor survival time observed in patients with breast cancer who have an increased expression of miR-711.
Figure 5.
miR-711 enhances the migration and invasion capabilities of breast cancer cells. (A) miR-711 overexpression promotes migration of human breast cancer MDA-MB-231 cells. The cells were transiently transfected with a miR-711 mimic or NC. The number of migrated cells was increased in cells transfected with miR-711 mimic compared with NC-transfected cells. (B) miR-711 knockdown reduces the migration of MDA-MB-231 cells. The cells were transiently transfected with a miR-711 inhibitor or NC, and the number of migrated cells was decreased in cells transfected with miR-711 inhibitor compared with NC-transfected cells. (C) miR-711 overexpression promotes invasion of MDA-MB-231 cells. The number of invading cells was increased in cells transfected with miR-711 mimic compared with NC-transfected cells. (D) miR-711 knockdown reduces the invasion of MDA-MB-231 cells. The number of invading cells was decreased in cells transfected with miR-711 inhibitor compared with NC-transfected cells. The number of migrated cells counted in 5 randomly selected fields of view (magnification, ×100) is presented as a histogram. miR-711, microRNA-711; NC, negative control.
Discussion
In recent years, mortality rates of breast cancer have continued to decrease due to advances in early diagnosis and treatment (2,3). However, thousands of women succumb to this disease each year worldwide. In clinical practice, positive outcomes and increased survival time have been proven to be associated with early diagnosis and precise therapy in breast cancer (1). Therefore, identification of effective prognostic biomarkers is urgently required to improve the survival time of patients with breast cancer. The dysregulation of miRNAs in breast cancer has been extensively investigated. In 2008, Gregory et al (28) revealed that the expression of the miR-200 family was abnormal in regions of metaplastic breast cancer tissues lacking E-cadherin. Wu et al (29) reported that miR-205 expression was significantly reduced in breast cancer tissues compared with matched normal breast tissues by using miRNA TaqMan PCR. Furthermore, it was observed that certain breast cancer cell lines, including MCF-7 and MDA-MB-231, expressed a decreased level of miR-205 compared with the non-malignant breast MCF-10A cell line (29). In addition, it was observed that ectopic expression of miR-205 significantly inhibited cell proliferation and growth, as well as cell invasion, and in animal models miR-205 suppressed lung metastasis (29). In 2012, Rivas et al (30) reported that miR-16 functioned as a tumor suppressor in breast cancer. It was demonstrated that miR-16 was involved in progestin-induced tumor growth, and overexpression of miR-16 repressed progestin-induced breast tumor growth in vitro and in vivo (30). Furthermore, it was demonstrated that miR-16 was significantly downregulated by hormonotherapy in an in vivo setting (30). These results suggested a novel mechanism of progestin-induced breast cancer growth that may have the potential to modulate a wide array of genes (30). Notably, it also demonstrated the involvement of miR-16 in HRG-induced breast cancer cell proliferation, confirming the ability of miR-16 to act as a tumor suppressor during breast cancer cell proliferation (30). To the best of our knowledge, there have been no studies that have investigated the role of miR-711 in human cancer. However, previous studies have investigated the role of miR-711 in organ injury and have reported miR-711 upregulation leading to various effects, including inhibition of collagen-1 expression in myocardial infarction and induction of neuronal cell death following traumatic brain injury (31).
The present study reports that miR-711 is aberrantly overexpressed in breast cancer tissues; this is similar to results observed in cutaneous T-cell lymphoma, in which miR-711 is overexpressed (32). Notably, the present study revealed that miR-711 overexpression is associated with poor OS and DFS times and is an independent prognostic factor in patients with breast cancer. Furthermore, the present in vitro experiments demonstrated that overexpression of miR-711 promotes proliferation, colony formation, migration and invasion of breast cancer cells, and that a knockdown of miR-711 significantly increases the percentage of apoptotic breast cancer cells. This indicates that miR-711 promotes the oncogenic proliferation of breast cancer cells by inhibiting the apoptosis of cells. Therefore, additional studies are required to investigate the target genes of miR-711 and the mechanism by which it functions in breast cancer cells.
In summary, the present study demonstrates for the first time, to the best of our knowledge, that miR-711 is an independent prognostic factor and plays an important oncogenic role in patients with breast cancer; however, the exact nature of this role remains to be determined. Thus, miR-711 may potentially be used as a biomarker for prognosis, and may be a future targeted therapy in patients with breast cancer.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China/Joint Research Fund for Oversea Scholar (grant no. 81228104) and the Research Fund of State Key Laboratory of Oncology in South China.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Autier P, Boniol M, La Vecchia C, Vatten L, Gavin A, Héry C, Heanue M. Disparities in breast cancer mortality trends between 30 European countries: Retrospective trend analysis of WHO mortality database. BMJ. 2010;341:(aug11 1): c3620. doi: 10.1136/bmj.c3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll R, Payne P, Waterhouse JAH, editors. Cancer Incidence in Five Continents. I. Geneva: Union Internationale Contre le Cancer; 1966. [DOI] [Google Scholar]
- 5.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 6.Saini KS, Taylor C, Ramirez AJ, Palmieri C, Gunnarsson U, Schmoll HJ, Dolci SM, Ghenne C, Metzger-Filho O, Skrzypski M, et al. Role of the multidisciplinary team in breast cancer management: Results from a large international survey involving 39 countries. Ann Oncol. 2012;23:853–859. doi: 10.1093/annonc/mdr352. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 9.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 11.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Yousef M, Showe L, Showe M. A study of microRNAs in silico and in vivo: Bioinformatics approaches to microRNA discovery and target identification. FEBS J. 2009;2762150:2156. doi: 10.1111/j.1742-4658.2009.06933.x. Liu CG, Calin GA, Volinia S and Croce CM: MicroRNA expression profiling using microarrays. Nat Protoc 3: 563-578, 2008. [DOI] [PubMed] [Google Scholar]
- 15.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 21.Cho WC. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao N, Yu H, Sun M, Zhang Y, Xu M, Gao W. MiRNA-711-SP1-collagen-I pathway is involved in the anti-fibrotic effect of pioglitazone in myocardial infarction. Sci China Life Sci. 2013;56:431–439. doi: 10.1007/s11427-013-4477-1. [DOI] [PubMed] [Google Scholar]
- 25.Greene FL, Page DL, Fleming ID, Fritz AG, editors. 6th. New York, NY: Springer Science+Business Media; 2002. American Joint Committee on Cancer: Cancer Staging Manual; pp. 221–240. [Google Scholar]
- 26.Körbler T, Grsković M, Dominis M, Antica M. A simple method for RNA isolation from formalin-fixed and paraffin-embedded lymphatic tissues. Exp Mol Pathol. 2003;74:336–340. doi: 10.1016/S0014-4800(03)00024-8. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas MA, Venturutti L, Huang YW, Schillaci R, Huang TH, Elizalde PV. Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res. 2012;14:R77. doi: 10.1186/bcr3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabirzhanov B, Stoica BA, Zhao Z, Loane DJ, Wu J, Dorsey SG, Faden AI. miR-711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death Differ. 2015 Oct 16; doi: 10.1038/cdd.2015.132. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralfkiaer U, Hagedorn PH, Bangsgaard N, Løvendorf MB, Ahler CB, Svensson L, Kopp KL, Vennegaard MT, Lauenborg B, Zibert JR, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL) Blood. 2011;118:5891–5900. doi: 10.1182/blood-2011-06-358382. [DOI] [PMC free article] [PubMed] [Google Scholar]