Table 2.
Structure and antiviral activity of K11777 analogs modified at the P3 position.
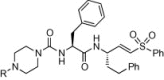 | |||||
|---|---|---|---|---|---|
| Compound | P3 substituent R= |
MW | pKaa | HIV-luc (SARS-CoV S)b IC50 (nM) |
HIV-luc (EBOV GP) IC50 (nM) |
| K11777 | Me | 575 | 7.02 | 0.32 ± 0.02 | 0.36 ± 0.02 |
| SMDC-256122 | Et | 589 | 7.29 | 0.04 ± 0.01 | 0.12 ± 0.01 |
| SMDC-256123 | i-Pr | 603 | 7.57 | 0.11 ± 0.01 | 0.25 ± 0.07 |
| SMDC-256157 | n-Pr | 603 | 7.59 | 0.24 ± 0.03 | 0.42 ± 0.03 |
| SMDC-256158 | Ph | 637 | 3.42 | 2.49 ± 0.34 | 2.69 ± 0.43 |
| SMDC-256159 | —CH2CH2OCH3 | 619 | 6.82 | 0.07 ± 0.02 | 0.16 ± 0.02 |
| SMDC-256160 | t-Bu | 617 | 7.87 | 0.08 ± 0.01 | 0.11 ± 0.03 |
| SMDC-256161 | Cyclopentyl | 629 | 8.01 | 0.25 ± 0.16 | 0.18 ± 0.01 |
| SMDC-256162 | Cyclopropylmethyl | 615 | 7.73 | 0.16 ± 0.03 | 0.10 ± 0.01 |
Calculated in MarvinSketch 5.5.0.1 from ChemAxon Ltd.
IC50 (inhibitory concentration) values of SARS-CoV or EBOV are the concentrations required to inhibit the infectivity of SARS-CoV or EBOV pseudotyped viruses on 293T-ACE2 cells by 50%, which were determined from dose response curves. A non-linear regression analysis based on the Sigmoidal dose response equation was applied to the percent inhibition and concentration data. Data is shown as means of quadruplicate measurements ± standard deviation. Values are representative of at least three independent experiments.
