Table 1. Methods used in the discovery and characterization of RNA modifications (2009-2014).
| Identity | Symbol | Structure | RNA Species | Discovery | Validation | Quantification |
|---|---|---|---|---|---|---|
| 5-Cyanomethyluridine | cnm5U |
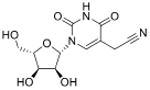
|
tRNA | Targeted analysis of mutant phenotype | Std, pMS3 | MRM |
| N6-Hydroxymethyladenosine | hm6A |
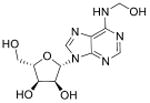
|
mRNA | Targeted analysis of modified synthetic ssRNA | Std, MS2 | MRM |
| N6-Formyladenosine | f6A |
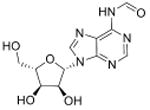
|
mRNA | Targeted analysis of modified synthetic ssRNA | Std, MS2 | MRM |
| Cyclic N6 Threonylcarbamoyl-adenosine | t6A |
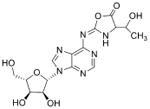
|
tRNA | Targeted analysis of natural product | Std, MSn, 18O-labeling, NMR | UV absorbance |
| 2-Geranylthiouridines* | ge5s2U |
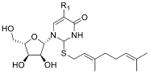
|
tRNA | LC-MS/MS screen of nucleotides | Std, MS2, 13C-labeling | External calibration |
| N6, N6-dimethyladenosine | m6,6A |
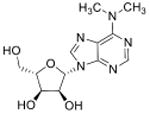
|
tRNA | LC-MS/MS screen of nucleotides | Std, pMS3 | MRM |
| 2,8-Dimethyladenosine | m2,8A |

|
rRNA | Targeted analysis of mutant phenotype | Std, MSn | Not performed |
| N-ribosylnicotinamide | r-NA |
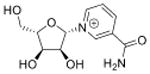
|
5′cap of sRNAs (>200 nt) | LC-MS/MS screen of nucleotides | Std, MS2, 13C-labeling | External calibration, 13C/15N-labeled isotope dilution |
| Co-enzyme A** | coA |

|
5′cap of sRNAs (>200 nt) | LC-MS/MS screen of nucleotides | Std, MS2, 18O-labeling | External calibration |
R1 can be hydrogen, methylaminomethyl- or carboxymethylaminomethyl- groups (Dumelin et al., 2012);
R2 can be hydrogen, succinyl-, acetyl- or methylmalonyl- groups (Kowtoniuk, Shen, Heemstra, Agarwal, & Liu, 2009); Std, synthetic standards; pMS3, pseudo-MS3 by controlled in-source fragmentation; MSn, tandem in-time fragmentation on ion-trap mass spectrometers.
