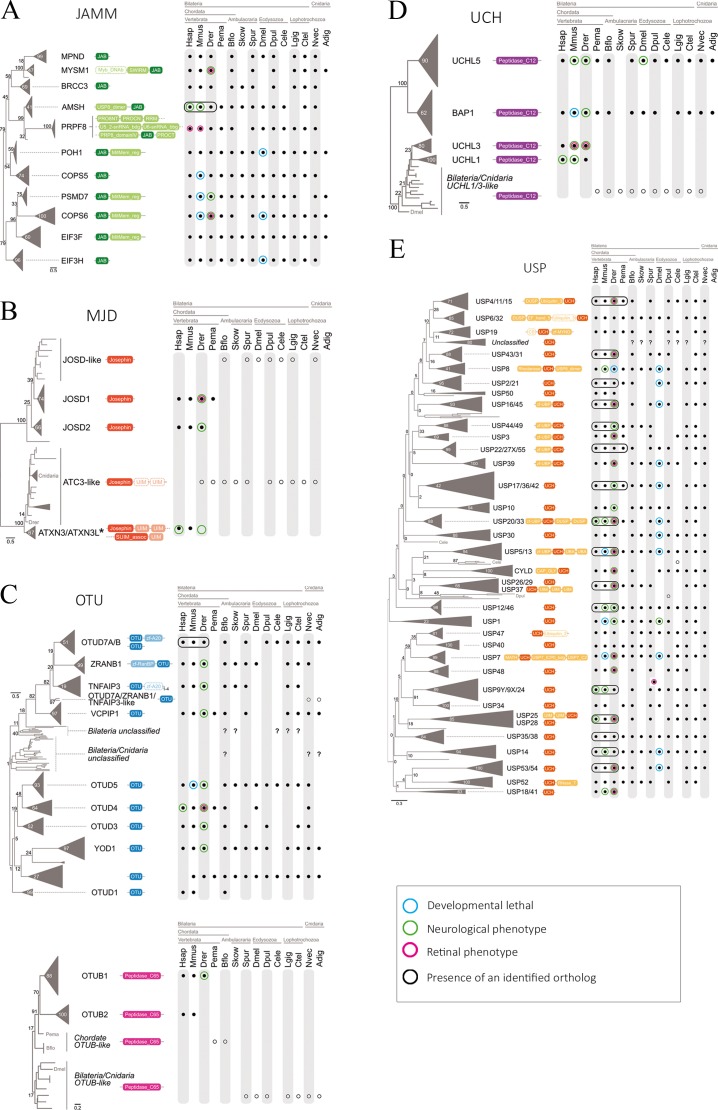
Fig 4. Phylogenetic analysis of DUB genes and neuronal/retinal phenotype.
Protein sequences from the catalytic region of each enzyme group were queried in complete genome sequences of 14 animal taxa and aligned. The protein domain architectures including the catalytic and accessory domain motifs are represented next to each DUB member (A, JAMM; B, MJD; C, OTU; D, UCH; and D, USP). Black dots indicate presence of the ortholog, whereas white dots indicate homologs that cannot be confidently assigned to a DUB type (see Results). Question marks represent statistically supported clades of uncharacterized DUBs. DUB sequences that are highly similar and cluster closely together appear collapsed under a common name. In general, invertebrates have a single representative member of the collapsed branch, whereas vertebrate genomes show one member of each paralog (species circled in black). Acropora digitifera USP homologs were excluded from the analysis as they impaired the resolution of the USP phylogeny. Genes reported to produce an abnormal neuronal phenotype when mutated are circled in magenta, whilst genes producing abnormal eye or retinal phenotype are circled in green. Genes whose mutation is lethal during developmental stages are circled in blue. An schematic summary of the DUB mRNA localization in the mouse retina (from ISH) is also presented next to the corresponding family. The intensity of the color indicates hybridization signal intensity. Retinal layers appear indicated as in Fig 2.