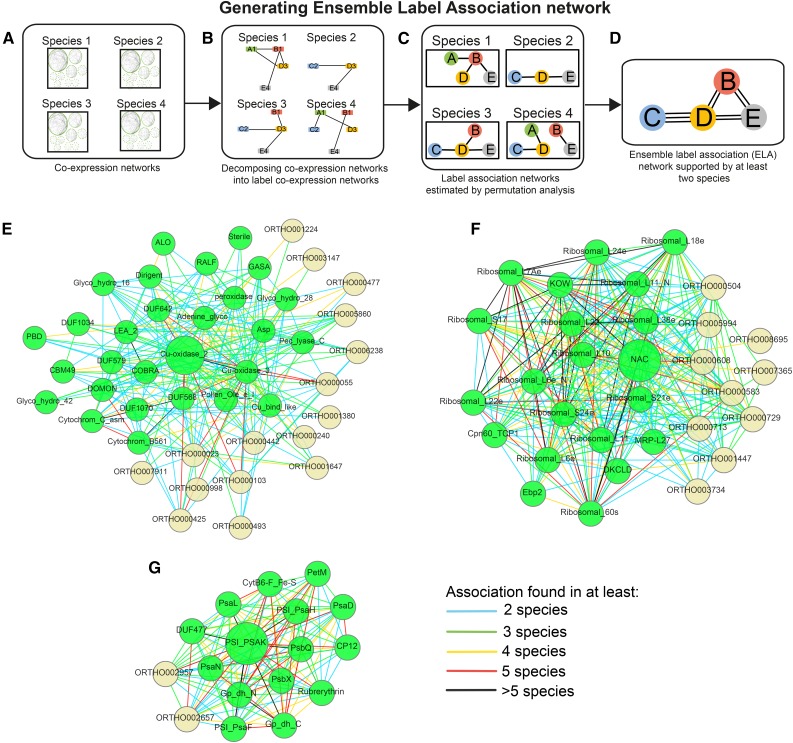
Figure 1.
Generating the Ensemble Label Association (ELA) network. A, Coexpression networks derived from the PlaNet platform are used as input. Each gene may be assigned multiple labels. B, The gene coexpression networks are decomposed into label coexpression networks, where nodes represent labels assigned to genes and edges represent coexpression relationships between the labels. C, Associations between labels are detected for each species by a label-based node cover and permutation test. The result is label association networks, where nodes represent labels and edges represent associations between the labels. D, Label association networks are combined into an ELA network. The number of edges (associations) that are conserved across the different species is determined. In this example, labels C and D are connected in three species (species 2, 3, and 4). E, ELA of Cu-oxidase_2. F, ELA of PSI_PSAK. G, ELA of NAC. Green and yellow nodes represent Pfam and PLAZA labels, respectively. Edges show in how many species an association was found.