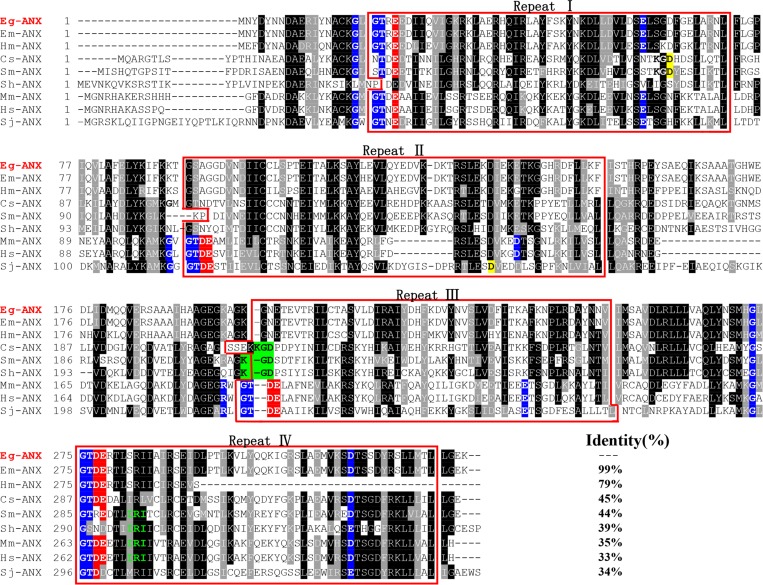
Figure 1.
Sequence alignment analysis of annexin B33 protein of Echinococcus granulosus (Eg-ANX) with homologous annexins. Alignment of the deduced amino acid sequence of E. granulosus annexin (GeneDB: EgrG_000041300) with homologs from other species. The following sequences were retrieved from the GenBank protein sequence database and aligned using the Clustal X program: Echinococcus multilocularis annexin (Em-ANX) (GenBank: CDI96708.1), Hymenolepis microstoma annexin (Hm-ANX) (GenBank: CDS34213.1), Clonorchis sinensis annexin (Cs-ANX) (GenBank: GAA40121.2), Schistosoma mansoni annexin (Sm-ANX) (GenBank: ACO90180.1), Schistosoma haematobium annexin (Sh-ANX) (GenBank: KGB36040.1), Mus musculus annexin (Mm-ANX) (GenBank: NP_081487.1), Homo sapiens annexin (Hs-ANX) (GenBank: CAG46637.1), and Schistosoma japonicum annexin (Sj-ANX) (GenBank: CAX70812.1). According to the Clustal X algorithm, regions of high identity and similarity between annexin sequences are shown as black and gray columns, respectively. The four repeat domains of annexin sequences are marked with red boxes. The actin-binding motifs (in repeat IV) are indicated by green letters. The type II Ca2+-binding sites are in white letters with blue backgrounds; the type III Ca2+-binding sites are in white letters with red and blue backgrounds. The KGD motifs are in black letters with green backgrounds. The percentage homology of Eg-ANX with other species is shown at the end of the alignments.