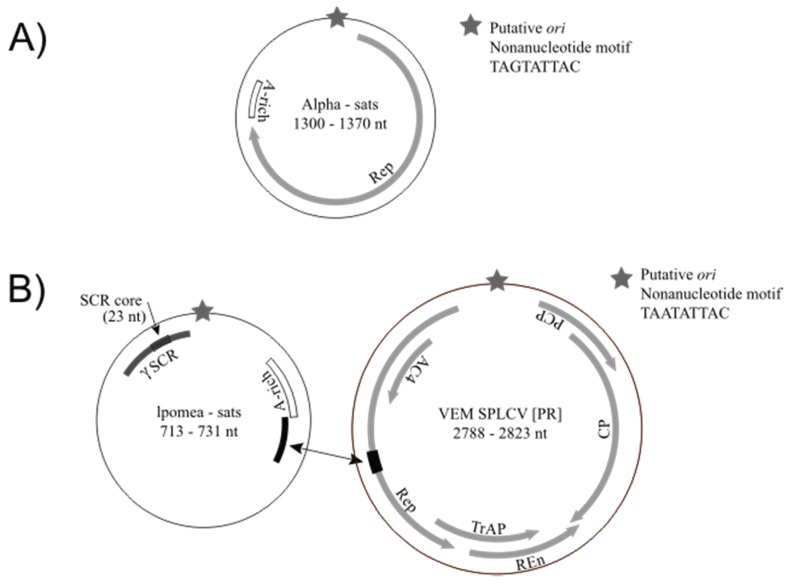
Figure 1.
Schematics depicting general genome organization and features observed in novel alphasatellites (A) and Ipomoea satellites (B) detected during this study. All genomes are characterized by a putative origin of replication (ori) marked by a stem-loop structure containing a conserved nonanucleotide motif. Alphasatellites encode a replication-associated protein (Rep) and exhibit an adenine-rich (A-rich) region. Ipomoea satellites do not exhibit any coding regions; however, these gammasatellites contain an A-rich region as well as a conserved region ~100 nt long shared among various gammasatellites (γSCR). The γSCR contains a 23 nt stretch where all gammasatellites reported to date share high similarities with betasatellites, named here the satellite common region (SCR) core. Some Ipomoea satellites from Puerto Rico share high identities with a 66 nt stretch (highlighted in black) found within the rep gene of sweet potato leaf curl virus (SPLCV) genomes detected in the same region.