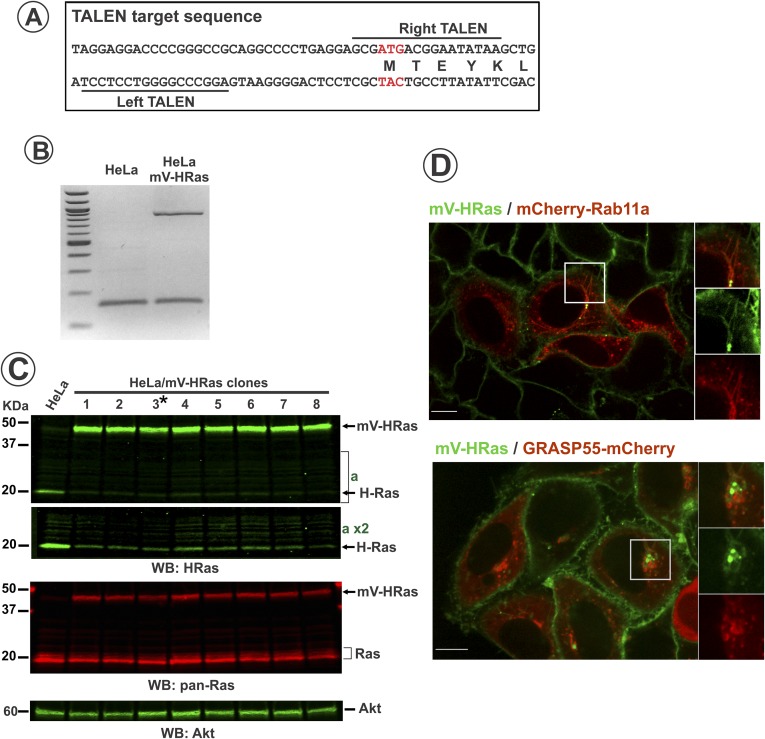
Fig. S2.
Characterization of mVenus-HRas in genome-edited HeLa cells. (A) TALEN target sequences in the HRAS gene. (B) PCR of 5′-UTR of HRAS gene from parental and edited HeLa cells. Note double band in HeLa/mVHRas sample confirms single allele edition. High–molecular-weight band corresponds to edited HRas allele. (C) Parental and several single-cell clones of HeLa/mV-HRas cells were lysed, and the lysates were probed by Western blotting with HRas, panRas, and Akt (loading control) antibodies. Asterisk shows the clone 3 used in experiments, although mVRas localization and dynamics were identical in all clones tested. Note increased (2.2-fold in clone 3 when normalized to loading control) expression of mV-Ras compared with HRas in parental cells, and reduced expression of untagged HRas in HeLa/mV-HRas cells (see part of blot “a” in high contrast), which is probably due to posttranslational control of the total expression level of HRas (mV-HRas plus untagged HRas). (D) HeLa/mVHRas cells were transfected with mCherry-Rab11 or GRASP55-mCherry. Images were acquired from living cells at 37 °C. Insets represent high-magnification images of each individual protein and a merge image of the region indicated by white rectangles. (Scale bars: 10 μm.)