Fig. 2.
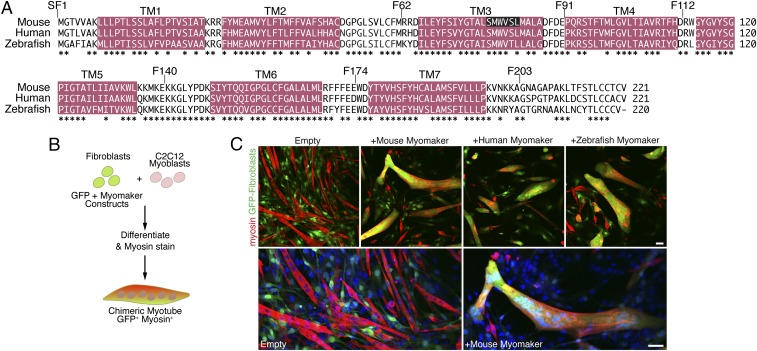
Myomaker orthologs are fusogenic. (A) Amino acid alignment of mouse, human, and zebrafish myomaker proteins. Magenta regions depict significant stretches of hydrophobic amino acids. TM refers to TM domains. Also shown are locations of FLAG epitopes preceding the indicated amino acid (SF1, F62, F91, F112, F140, F174, and F203). Residues deleted in the CRISPR/Cas9 myomaker KO clone 1B5 are displayed with white font and black background. *Sequence conservation. (B) Schematic showing the heterologous cell–cell fusion system. Fibroblasts were infected with GFP and myomaker constructs and mixed with C2C12 myoblasts. The cultures were differentiated for 4 d and then immunostained with myosin antibody as a marker for myocytes. GFP-positive (fibroblast origin), myosin-positive (myoblast origin) chimeric myotubes, in yellow/orange, indicate fusion between the two cell populations. (C) Fibroblasts infected with control (empty) and GFP viruses do not fuse to C2C12 cells. Expression of mouse, human, or zebrafish myomaker in fibroblasts promotes dramatic fusion with muscle. Lower shows enlarged images of cells infected with empty and mouse myomaker retroviruses with Hoechst-stained nuclei. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.)