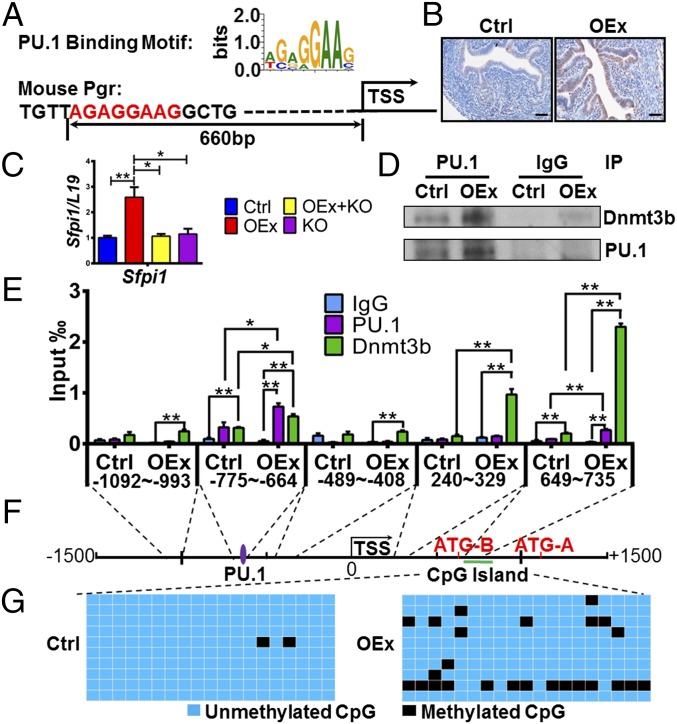
Fig. 4.
DNA hypermethylation of the Pgr gene mediated by PU.1/Dnmt3b. (A) Predicted PU.1 binding site on the Pgr promoter. PU.1 is up-regulated in N1ICD OEx mice by (B) immunohistochemistry and (C) quantitative PCR; quantitative expression levels of PU.1 (immunohistochemistry) are shown in Fig. S3C. (D) The PU.1 and Dnmt3b interaction is stronger in OEx mice than that in control mice (n = 3). (E) Binding of PU.1 and Dnmt3b protein on the Pgr promoter was detected by ChIP (n = 3). Binding efficiency of both PU.1 and Dnmt3b is significantly higher in OEx mice than control mice at positions of the predicted PU.1 motif (−755 to −644) and the CpG island (649–735). (F) Relative positions of the PU.1 binding site, the Pgr transcription start site, and the CpG island. (G) The number of methylated CpGs (black cells) is much higher in OEx mice than in control mice. Blue cells are unmethylated CpGs. All data are collected at 3.5 dpc. Ctrl, control; IP, immunoprecipitation. *P < 0.05; **P < 0.01. (Scale bar: 100 µm.)