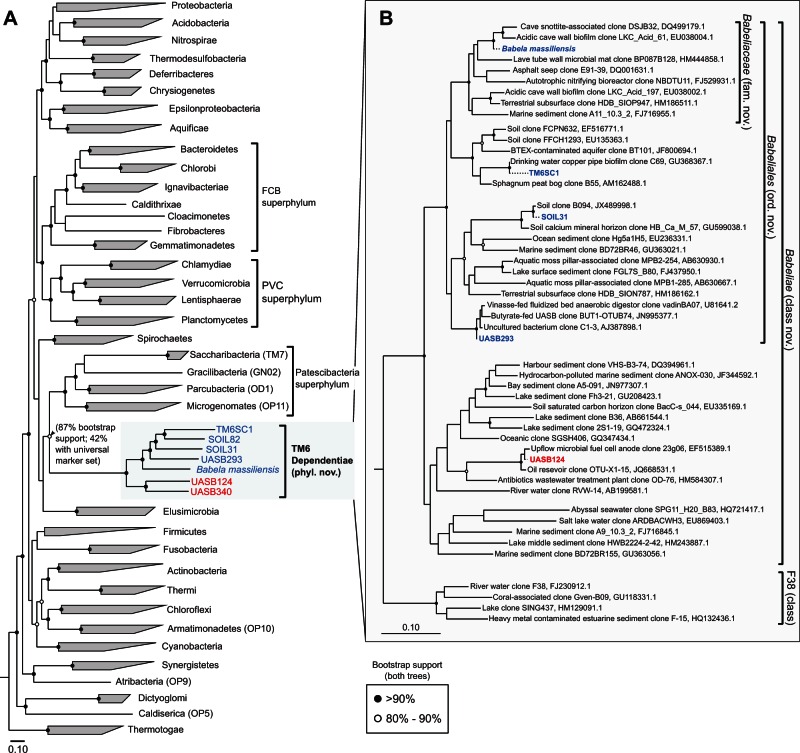
Fig. 1.
Phylogenetic relationships of candidate phylum TM6 bacteria. (A) Genome-based phylogeny of TM6 bacteria relative to other bacterial phyla. A maximum-likelihood tree was constructed from 2,175 bacterial reference genomes using an amino acid alignment of 83 phylogenetically informative bacterial marker genes. Candidatus Acetothermus autotrophicum (OP1) was used as the outgroup (Takami et al. 2012). (B) 16S rRNA gene-based phylogeny of TM6 bacteria using 16S sequences obtained from population genomes in this study (dotted lines) and representative sequences from the Greengenes database. The population genome labels for TM6 bacteria in both trees are highlighted in blue or red to reflect order-level relatedness. In both trees, black circles at nodes indicate >90% bootstrap support and open circles indicate 80–90% bootstrap support. The scale bars indicate 0.1 amino acid and nucleotide substitutions per site for panels (A) and (B), respectively.