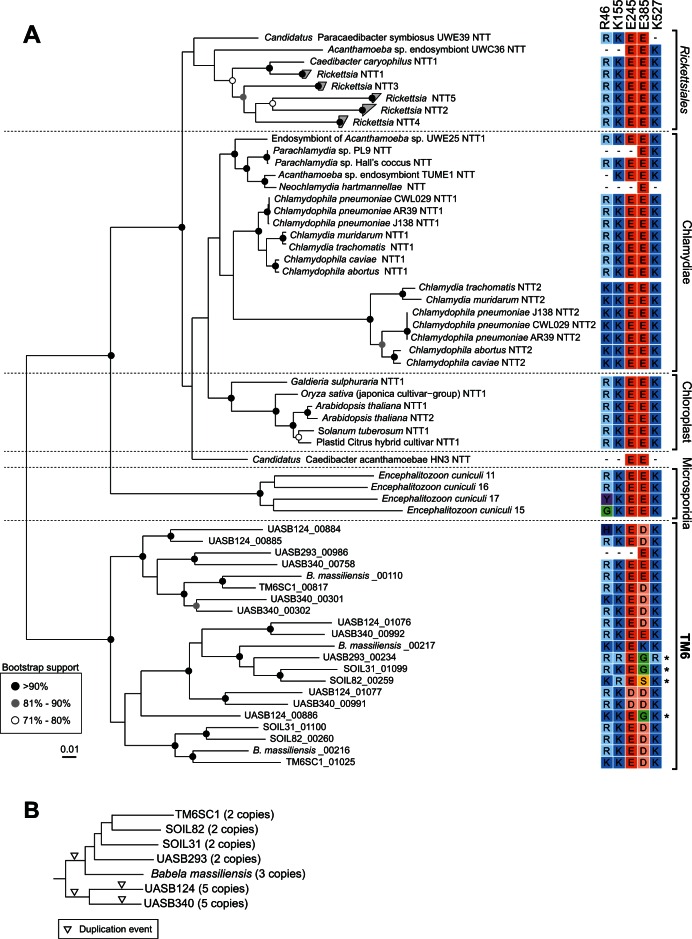
Fig. 4.
Phylogenetic relationships of TM6 ATP/ADP translocase proteins. (A) Unrooted maximum-likelihood tree of aligned ATP/ADP transporter (nucleotide transporter; NTT) amino acid sequences identified in the TM6 genomes and representatives of the Chlamydiae, Rickettsiales, chloroplasts, and Encephalitozoon cuniculi (microsporidia; Schmitz-Esser et al. 2004). Paralogs are indicated as NTT1–NTT5. Five-digit numbers included in the TM6 bacteria labels represent sequential gene ordering (i.e., relative genomic location) in the respective population genomes as annotated by PROKKA. The scale bar indicates amino acid substitutions per site. Alignment of key ATP/ADP translocase amino acid residues for transport efficiency and substrate specificity is depicted in a colored matrix adjacent to the tree, in which conservative amino acid mutations are represented by shades of blue (H, K, and R) or orange (D and E). Asterisks indicate paralogs with substitutions in these key residues. Residue position R46 is based on the Rickettsia prowazekii ATP/ADP translocase (Alexeyev and Winkler 2000) whereas positions K155, E245, E385, and K527 are based on the Arabidopsis thaliana ATP/ADP translocase (Trentmann et al. 2000). (B) Genome-based tree of the TM6 phylum (inset from fig. 1A) showing inferred ATP/ADP translocase gene duplication events and number of paralogs identified in the respective population genomes. Inverted triangles indicate gene duplications that can be inferred from ATP/ADP translocase paralog count, genomic location, and gene phylogeny (fig. 4A).