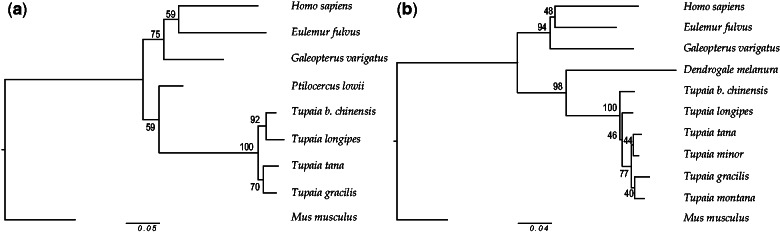
Fig. 3.
Phylogenies of Euarchonta based on intron (a) and synonymous (b) sites of the OPN1SW. The evolutionary history was inferred by using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Felsenstein 1985). The evolutionary distances are in units of the number of differences per site (Nei-Gojobori 1986). The analysis involved 9 (a) and 11 (b) nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 774 (a) and 354 (b) positions in the final data set. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013).