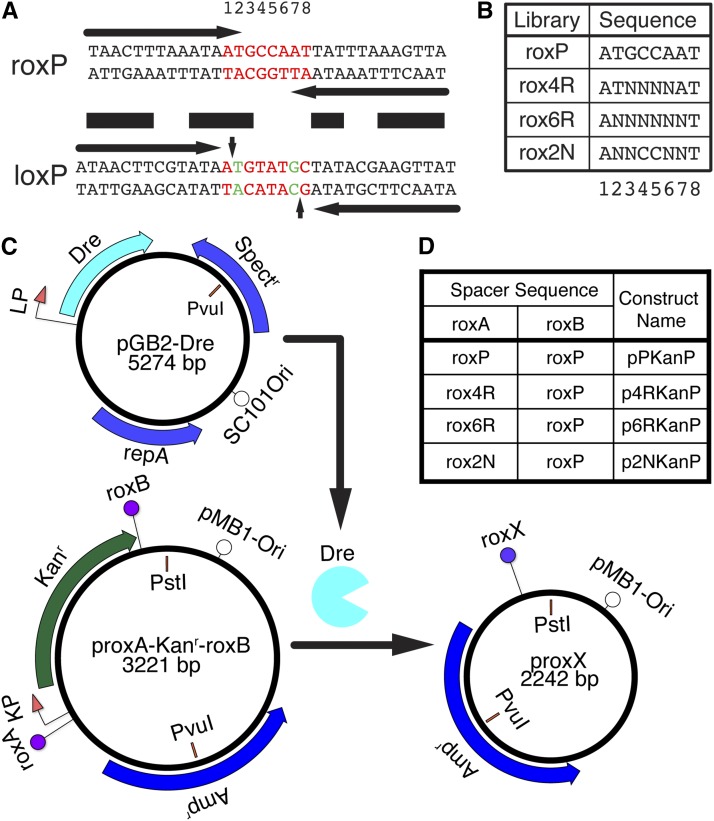
Figure 1.
Random nucleotide libraries targeted at the roxP spacer region. (A) Comparison of roxP (top) and loxP (bottom) wild type sites. Horizontal arrows mark the inverted repeats for each target site and the black horizontal bars label identical base pairs. Regions highlighted in red correspond to the spacer region of the loxP site and the equivalent base pairs on the roxP site (numbered 1–8). The vertical arrowheads on the loxP sequence mark the sites of catalytic attack and strand break during Cre recombination. Mutations at positions 2 and 7 in the loxP spacer, highlighted in green, are used in the lox2272 mutant. (B) Libraries carrying random nucleotides indicated by N’s inserted at positions 3–6 (rox4R), 2–7 (rox6R) and 2, 3, 6, and 7 (rox2N) in the spacer region of roxP. (C) Plasmids and recombination test used in this paper. pGB2-Dre contains a replication cassette consisting of the repA protein and SC101 origin of replication (ensuring 1–5 copies/cell), a spectinomycin resistance gene (Spcr), and a Dre open reading frame driven by a lac promoter (LP). proxA-Kanr-roxB contains a pMB1 origin (generating >75 copies/cell), an ampicillin resistance gene, and a kanamycin (Kanr) resistance cassette, flanked by two rox sites arranged in direct orientation, roxA and roxB. If both rox sites are occupied by wild type roxP sequences (pPKanP vector), Dre recombination results in deletion of the intervening fragment containing the Kanr element. In the unrecombined plasmid, PvuI + PstI digestion results in 1.8 kb and 1.4 kb fragments, while after Dre recombination and Kanr removal the 1.8 kb fragment is reduced to 0.8 kb, while the 1.4 kb fragment (containing the pMB1-Ori) is unaffected. (D) Wild type roxP sites were inserted at the roxB position, while the mutant rox sites carrying the random nucleotide libraries shown in B were cloned into the roxA position, generating the p4RKanP, p6RKanP, and p2NKanP vectors.