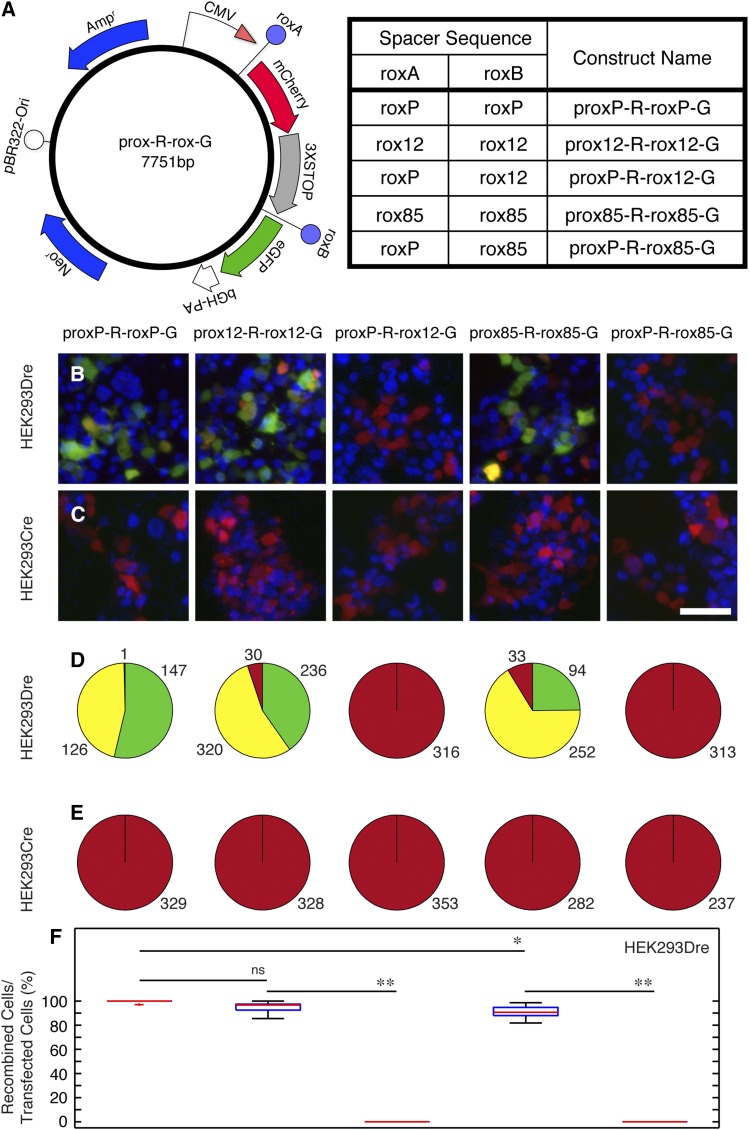
Figure 6.
Rox mutants rox12 and rox85 recombine with themselves but not with roxP in eukaryotic cells. (A) In the prox-R-rox-G eukaryotic expression vector, a CMV promoter is driving transcription of mCherry before Dre recombination between the roxA and roxB sites, and of eGFP after recombination. rox12 and rox85 mutants or roxP wild type control were placed at both roxA and roxB sites to generate prox12-R-rox12-G, prox85-R-roxP-85 and proxP-R-roxP-G vectors, suitable for testing the ability of each rox site to recombine with itself. In addition, rox12 or rox85 mutants were cloned at the roxB site in combination with a roxP at the roxA site, to generate proxP-R-rox12-G and proxP-R-rox85-G, to test the compatibility between rox mutants and roxP wild type. (B, C) HEK293 cells stably expressing either the Dre (B) or Cre (C) recombinase were transfected with the constructs described in A. (D, E) Numbers in pie charts indicate the sums for red (mCherry only), green (eGFP only), and yellow (double positive) cells over six individual 20 × fields derived from two distinct transfected coverslips for each of the experimental conditions in B and C. No eGFP positive cells were seen in HEK293Dre cells transfected with proxP-R-rox12-G and proxP-R-rox85-G (B, D, columns 3 and 5) or HEK293Cre cells transfected with either one of the five constructs (C, E). (F) Box-whisker plots quantitating the percent of recombination positive cells in the HEK293Dre experiments. The percentage represents 100 × (eGFP+ + eGFP+mCherry+)/(eGFP+ + eGFP+mCherry+ + mCherry+). Statistical significance was determined by student t-test and Kolmogorov–Smirnov test (n.s. = P > 0.05, * = P < 0.05, ** = P < 0.005). Scale bar in C = 50 µm.