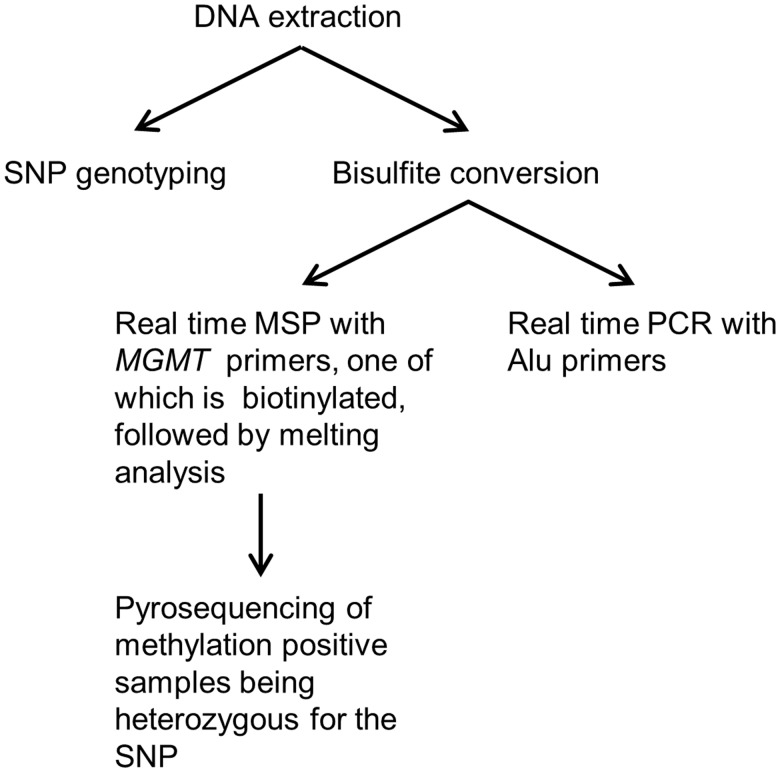
FIGURE 1.
Overview of the quantitative methylation-specific PCR (qMSP)-pyrosequencing workflow. First, genomic DNA is used for genotyping the single nucleotide polymorphism (SNP) in the region between the MSP primers. Then, for the methylation analysis, genomic DNA is bisulfite converted. Next, 2 separate real-time PCRs are performed; one with MSP primers (one of which is biotinylated) targeting the region of interest, and one with primers capable of amplifying both methylated and unmethylated templates targeting a control region (Alu sequences depleted of CpG sites). The data obtained are used to calculate the level of methylation in the samples relative to in vitro methylated DNA. Melting analysis is performed as an integrated part of the real-time PCR runs to confirm that the observed amplification is specific. Subsequently, amplification products are pyrosequenced to allow allelic methylation analysis for the patients being heterozygous for the SNP found between the MSP primers.