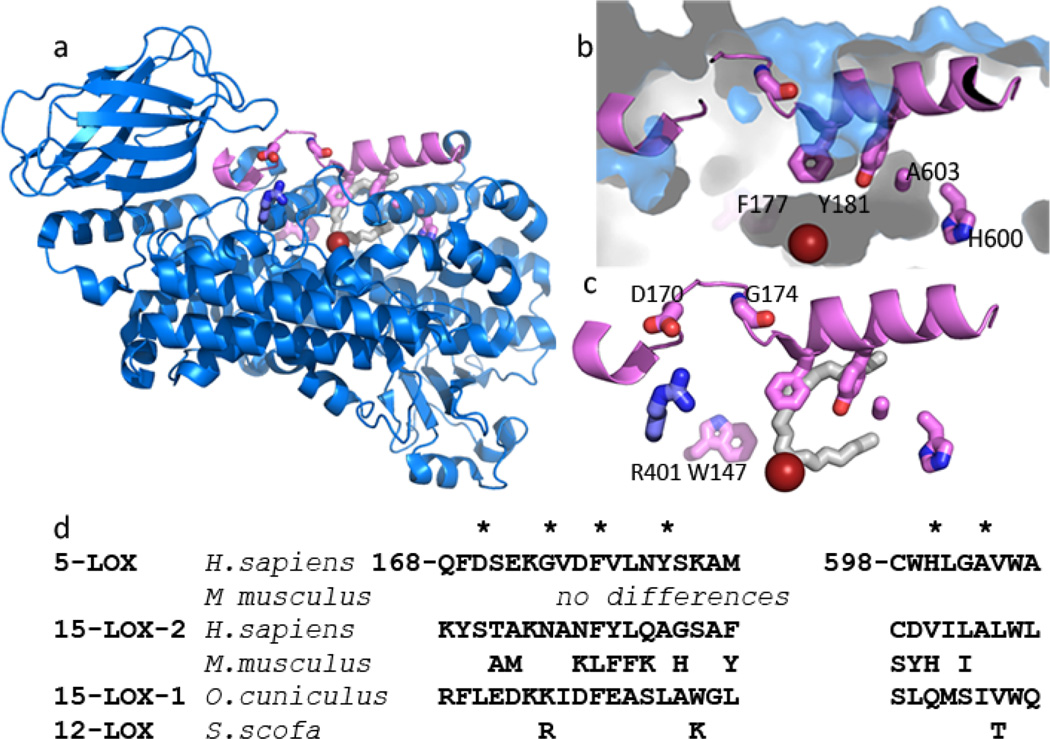
Fig. 2. St-5-lipoxygenase has an encapsulated active site.
(a) Cartoon rendering of St-5-lipoxygenase (3O8Y). The region corresponding to helix α2 is in pink. The iron is shown as an orange sphere. The amino acids mutated in this study are in stick rendering (C pink, O red, N blue). The white stick rendering marks the position of the substrate mimic (C8E4) in the homologue 15-LOX-2. (b) Close-up of the internal cavity with the mutated amino acids in stick rendering. The proximity of A603 and Y181 is visible in this view. (c) D170 and G174 are 5-LOX specific amino acids that appear to confer its unique conformation of α2. D170 is positioned to participate in a salt-link with the conserved R401. The inhibitor (C8E4) in the 15-LOX-2 structure is superimposed in white stick rendering to indicate where the substrate mimic is bound in that enzyme. (d) Sequences of the mutated regions from related lipoxygenases. The positions mutated are marked with *.