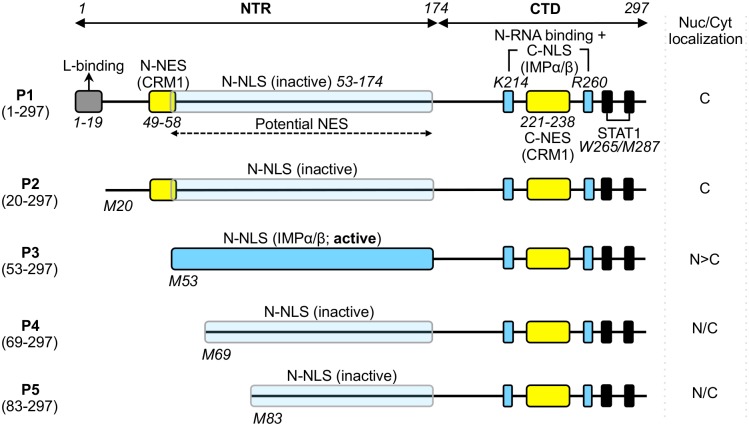
Fig 1. Domain structure of RABV P-protein.
P-protein is shown schematically with key domains/sequences indicated; residue positions are indicated by italicized numbering. The RABV P gene encodes full length P1 (residues 1–297) and N-terminally truncated isoforms P2-P5 (expressed via ribosomal leaky scanning that initiates translation from internal in-frame AUG codons corresponding to methionines M20, M53, M69 and M83 of P1 [13]). P1 alone contains residues 1–19 that are required for association with the viral L-protein so that P1 can act as the polymerase cofactor [18]. All P-protein isoforms contain the CTD which incorporates the binding sites for viral genome–associated N-protein (N-RNA), [17,23–25] and STAT1 (black boxes) [11,19,26]. P-protein interaction with EXPs and IMPs is mediated through two CRM-1-binding NESs (yellow boxes) [27,28] and two IMPα/β-binding NLSs (blue boxes, described elsewhere [9,27] and in this study), located in both the NTR and CTD. A third potential NES has been suggested to exist within NTR residues 53–174 [9]. The NTR-localized N-NLS is activated in P3 due to truncation of residues 1–52, which also truncates/deactivates the N-NES [9,27]. The P-CTD localized C-NLS, which includes key residues K214/R260 (blue boxes) of a positively charged patch on the surface of the CTD, is characterized in this study; the positive patch has also been implicated in binding to N-RNA [19,24,29–31], suggesting that the N-RNA-binding site overlaps with the C-NLS [24,25,27,28]. The nucleocytoplasmic (Nuc/Cyt) localisation of each isoform is indicated (cytoplasmic (C), nuclear (N), diffuse (N/C)).