Fig 7. Binding of NRSF proteins within the Δ8 mariner DNA segments.
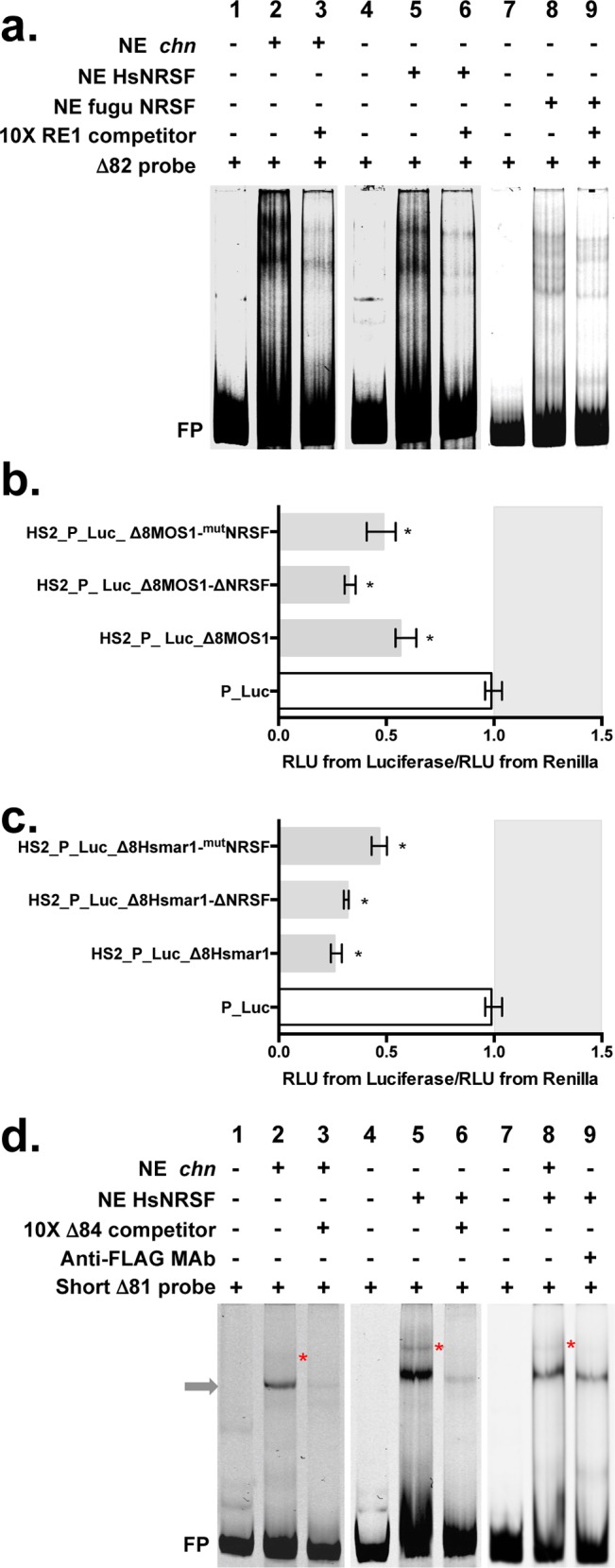
(a) DNA binding of chn, HsNRSF, fuguNRSF to the Δ82 segment of MOS1 (10 pmole of ATTO-labelled probe/lane). Lanes 1, 4 and 7 correspond to probe controls. Lanes 2, 5 and 8 show shifted complexes with each of the three NE. The specificity of the shifted complexes observed in lanes 2, 5 and 8 was verified by adding 10X of unlabelled RE1 DNA segments (lanes 3, 6 and 9). These experiments were done in triplicate and representative pictures are shown. (b) and (c) Expression of the Firefly and the Renilla luciferase marker genes using transient expression assays in HeLa cells. The assays were performed with three variants of Δ8-MOS1 in c and Δ8-HSMAR1 in d. The sequence of these variants is supplied in S11 Fig. Each histogram bar corresponds to the median value obtained from three experiments done in triplicate. Bars correspond to quartiles 1 and 3. The median ratios RLU from Firefly/RLU from Renilla were calculated as indicated in Fig 2. The area where the ratios “RLU from Firefly/RLU from Renilla” were above 1 (i.e. where no silencer effect is observed) is coloured in grey. * indicates a significant difference (p<0.05) with the P_Luc controls. (d) DNA binding of chn and HsNRSF to a shortest version of the Δ81 segment of MOS1 (10 pmole of ATTO-labelled probe/lane; Sequence supplied in S4 Fig). Lanes 1, 4 and 7 correspond to probe controls. Lanes 2, 5 and 8 show shifted complexes with each of the two NEs. The grey arrow locates a complex resulting from the binding of YY1, as shown in Fig 6D. The specificity of this YY1 complex in lanes 2 and 5 was verified by adding 10X of unlabelled Δ84 DNA segment that contains two YY1 binding sites. The red stars in lanes 2, 5 and 8 locate a complex that is absent when HeLa NE (Fig 6D) are used. The involvement of HsNRSF in this second complex is shown in lane 9, using a specific antibody that leads to its destabilization. It should be noted that this second complex is also sensitive to competition with the unlabelled Δ84 DNA segment (lanes 3 and 6).
