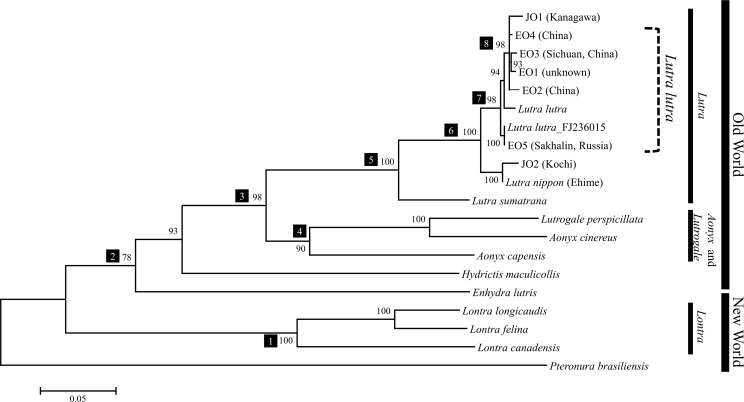
Fig 3. Phylogenetic tree for Lutrinae based on the partial mtDNA together with the L. nippon (Ehime).
This ML tree was based on the partial ND5 gene (692 bp) and complete cytb gene (1,134 bp) dataset, which included the L. nippon (Ehime) sequence (224 bp) [10]. This tree was estimated using the GTR+Γ+I model. Numbered boxes denote nodes. The nodal number indicates the BP value. BP was estimated on the basis of 1,000 bootstrap replicates. The evolutionary constraints on the nucleotide substitutions must differ between the first, second, and third codon positions; therefore, we specified partitions for each region. OTUs without localities are previously reported data (Koepfli and Wayne [1]; Koepfli et al. [12]). Data for L. lutra (South Korea) is FJ236015.