Figure 3.
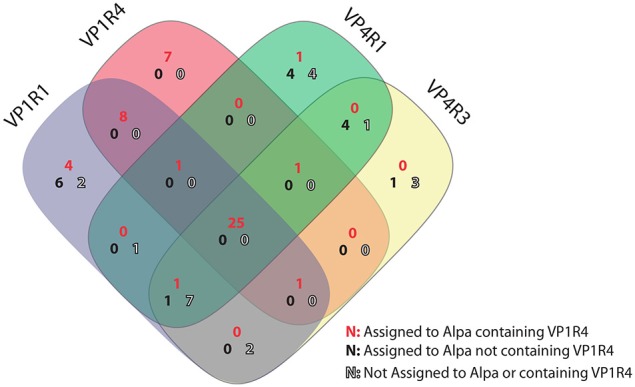
Consistency among HMM seeds. Venn diagram representing shared contigs reconstructed by progressive assembly using GenSeed-HMM with profile HMM seeds VP1R1, VP1R4, VP4R1, and VP4R3. Contigs were included in the same cluster when presenting at least 90% similarity at the nucleotide level covering at least 90% of the length of the shortest contig. Contigs were then taxonomically classified by blastx to reference proteins from Microviridae and searched for the presence of the VP1R4 seed using hmmsearch. A large percent of shared contigs among all four seeds is observed and belonging to Alpavirinae genomes covering the VP1R4 seed. Notice that contigs that were not present within the VP1R4 seed were usually not assigned to Alpavirinae (low precision) or do not contain the VP1R4 region, suggesting potential shorter non-overlapping contigs.
