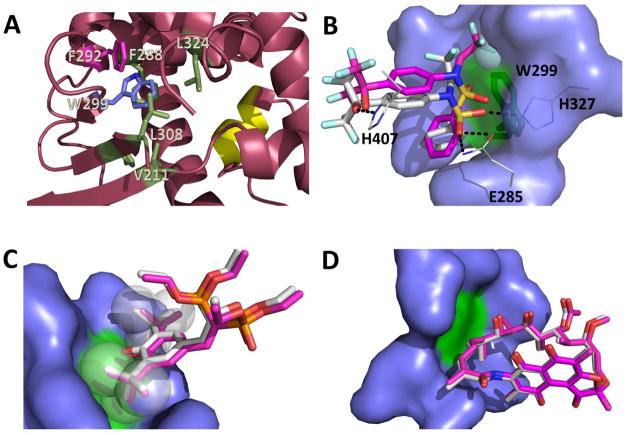
Figure 7. Effects of W299 mutation on hPXR activity as explained by computational analysis of hPXR structures.
(A) W299 (blue stick) contacts several nonpolar residues (green and pink sticks) forming an important hydrophobic patch in the hPXR-LBD for ligand binding and structural integrity, which can be disrupted by replacement of W299 with charged residues (pdb code 4XHD). F292 (pink stick) is involved in π-stacking contact with W299. The AF-2 helix is represented in yellow. (B) TO docked to the W299A mutant (carbon atoms in pink) in relation to TO as observed in the WT crystal structure (carbon atoms in white). Hydrogen bonds to protein residues (lines with carbon atoms in white) are indicated with dashes. The sphere represents the region in the TO molecule to most likely clash against W299 (green stick) (C) SR docked to the W299A mutant (carbon atoms in pink) in relation to SR as observed in the WT crystal structure (carbon atoms in white). The t-butyl groups of the ligand (spheres) prevent the ligand from moving around due to steric hindrance even after additional space is created by replacement of W299 (green) with alanine. (D) RIF docked to the W299A mutant (carbon atoms in pink) in relation to RIF as observed in the WT crystal structure (carbon atoms in white). The large molecule RIF cannot move around even when additional space is created following W299 (green) replacement by alanine. The amino acid residues that were made flexible during docking are represented as blue surface (B–D).